Fornix
In this script, you can find the results of the analysis across the fornix. To examine individual differences, we used K-means clustering in an exploratory analysis:
K-means clustering was conducted based on the total level of brain connectivity (total; FA at imaging timepoint one plus timepoint two: higher FA total level implies greater connectivity, lower FA total levels imply lower connectivity) and the development of brain connectivity from imaging timepoint one to two (trajectory; FA at timepoint two minus timepoint one).
Multinomial logistic regression was used to predict cluster membership based on adversity experiences at baseline. To adjust for multiple comparisons, p-values were Bonferroni-corrected in the actual paper. We corrected for the number of brain regions (four) and adversity scales (five).
Total & Trajectory
K-means Clustering
set.seed(1225)
Kmeans_Fornix <- kmeans(AdvDTI[,19:20], 3, iter.max = 10000, nstart = 50, algorithm="Lloyd") Groups <- as.factor(Kmeans_Fornix$cluster)
samplesize <- 10808
id=factor(1:samplesize)
PlotCluster <- data.frame(c(AdvDTI$Fornix_FA_t1), #select FA at time 1
c(AdvDTI$Fornix_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(Groups)) #insert cluster number
colnames(PlotCluster)<-c('T1','T2','ID','Cluster') #give column name
PlotAllClusters<-melt(PlotCluster,by='ID')
ggplot(PlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2","3")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2", "3" = "Cluster 3")))+
scale_color_manual(values = my_colors,
breaks =c("1","2","3"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))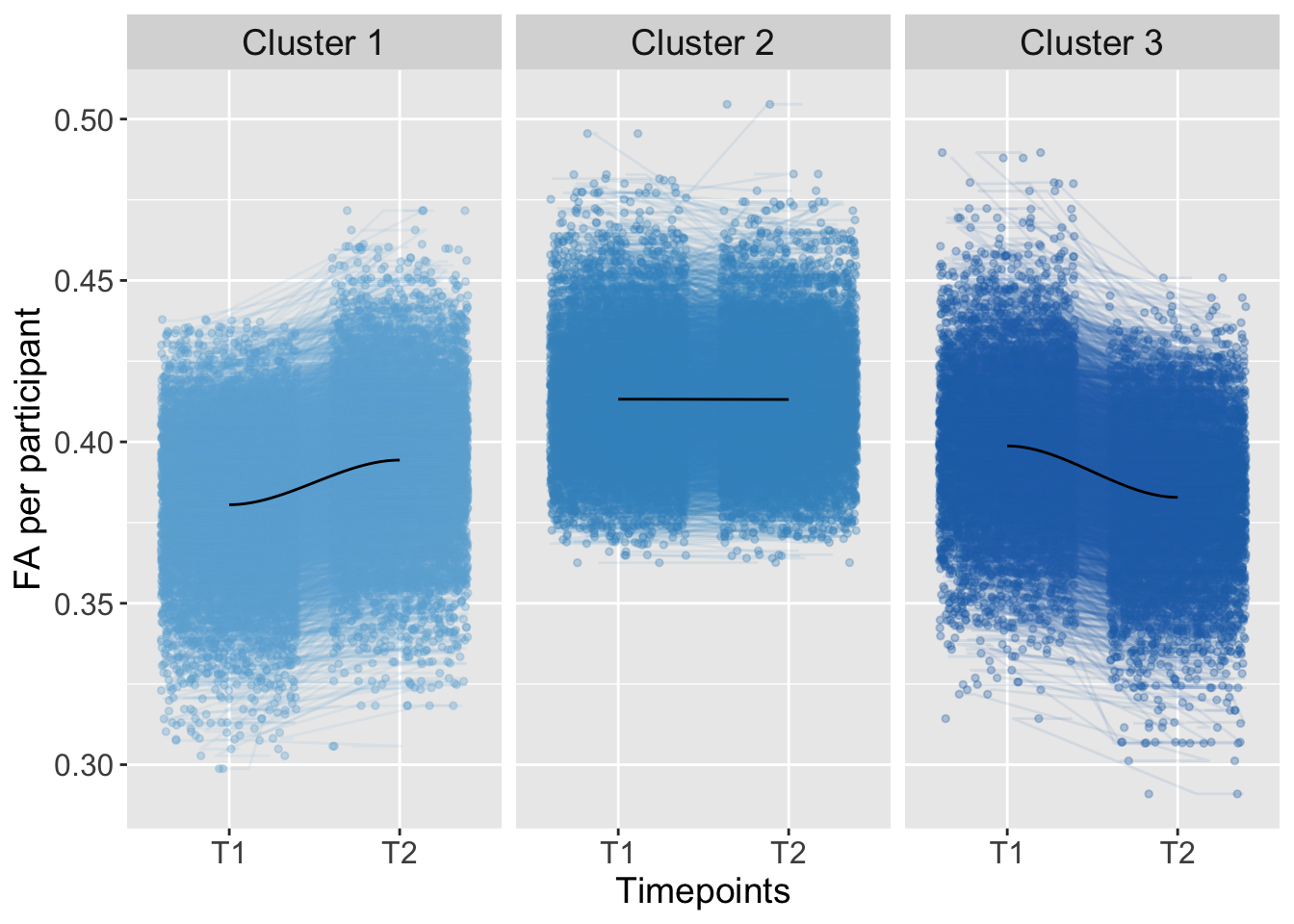
Regression results
AdvDTI$Groups <- as.factor(Kmeans_Fornix$cluster)
AdvDTI[,2:20] <- apply(AdvDTI[,2:20],2, function(x) as.numeric(as.character(x)))
mod_PTSD_Fornix = multinom(Groups ~ PTSDSum, data=AdvDTI)
mod_School_Fornix = multinom(Groups ~ SchoolSum, data=AdvDTI)
mod_Family_Fornix = multinom(Groups ~ FamilySum, data=AdvDTI)
mod_Safety_Fornix = multinom(Groups ~ Safety, data=AdvDTI)
mod_SES_Fornix = multinom(Groups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_Fornix, type=3) ## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 2.7911 2 0.2477summary(mod_PTSD_Fornix)## Call:
## multinom(formula = Groups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) PTSDSum
## 2 -0.07936731 0.006820816
## 3 -0.22215855 0.037950699
##
## Std. Errors:
## (Intercept) PTSDSum
## 2 0.02291514 0.02390163
## 3 0.02381277 0.02393429
##
## Residual Deviance: 23655.71
## AIC: 23663.71zPTFornix <- summary(mod_PTSD_Fornix)$coefficients/summary(mod_PTSD_Fornix)$standard.errors
zPTFornix## (Intercept) PTSDSum
## 2 -3.463531 0.2853703
## 3 -9.329387 1.5856206pPTFornix <- (1 - pnorm(abs(zPTFornix), 0, 1)) * 2
pPTFornix## (Intercept) PTSDSum
## 2 0.0005331347 0.7753605
## 3 0.0000000000 0.1128254Anova(mod_School_Fornix, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 2.6657 2 0.2637summary(mod_School_Fornix)## Call:
## multinom(formula = Groups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) SchoolSum
## 2 -0.07972975 -0.03732207
## 3 -0.22201065 -0.01268107
##
## Std. Errors:
## (Intercept) SchoolSum
## 2 0.02291772 0.02312102
## 3 0.02380911 0.02389804
##
## Residual Deviance: 23655.84
## AIC: 23663.84zSFornix <- summary(mod_School_Fornix)$coefficients/summary(mod_School_Fornix)$standard.errors
zSFornix## (Intercept) SchoolSum
## 2 -3.478956 -1.6142051
## 3 -9.324611 -0.5306322pSFornix <- (1 - pnorm(abs(zSFornix), 0, 1)) * 2
pSFornix## (Intercept) SchoolSum
## 2 0.0005033718 0.1064829
## 3 0.0000000000 0.5956737Anova(mod_Family_Fornix, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## FamilySum 10.892 2 0.004314 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Family_Fornix)## Call:
## multinom(formula = Groups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## (Intercept) FamilySum
## 2 -0.07900797 0.07578300
## 3 -0.22115141 0.03578014
##
## Std. Errors:
## (Intercept) FamilySum
## 2 0.02292968 0.02297718
## 3 0.02381918 0.02407378
##
## Residual Deviance: 23647.61
## AIC: 23655.61zFFornix <- summary(mod_Family_Fornix)$coefficients/summary(mod_Family_Fornix)$standard.errors
zFFornix## (Intercept) FamilySum
## 2 -3.445663 3.298185
## 3 -9.284593 1.486270pFFornix <- (1 - pnorm(abs(zFFornix), 0, 1)) * 2
pFFornix## (Intercept) FamilySum
## 2 0.0005696594 0.000973121
## 3 0.0000000000 0.137207722Anova(mod_Safety_Fornix, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## Safety 15.432 2 0.0004456 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Safety_Fornix)## Call:
## multinom(formula = Groups ~ Safety, data = AdvDTI)
##
## Coefficients:
## (Intercept) Safety
## 2 -0.07928747 0.00596028
## 3 -0.22302006 0.08535376
##
## Std. Errors:
## (Intercept) Safety
## 2 0.02292245 0.02337139
## 3 0.02383249 0.02378387
##
## Residual Deviance: 23643.07
## AIC: 23651.07zSafFornix <- summary(mod_Safety_Fornix)$coefficients/summary(mod_Safety_Fornix)$standard.errors
zSafFornix## (Intercept) Safety
## 2 -3.458943 0.2550246
## 3 -9.357814 3.5887242pSafFornix <- (1 - pnorm(abs(zSafFornix), 0, 1)) * 2
pSafFornix## (Intercept) Safety
## 2 0.0005422988 0.7987040922
## 3 0.0000000000 0.0003323001Anova(mod_SES_Fornix, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## SES 4.7417 2 0.0934 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_SES_Fornix)## Call:
## multinom(formula = Groups ~ SES, data = AdvDTI)
##
## Coefficients:
## (Intercept) SES
## 2 -0.07935304 0.004094055
## 3 -0.22191407 0.047845901
##
## Std. Errors:
## (Intercept) SES
## 2 0.02292032 0.02329295
## 3 0.02381516 0.02386817
##
## Residual Deviance: 23653.76
## AIC: 23661.76zSESFornix <- summary(mod_SES_Fornix)$coefficients/summary(mod_SES_Fornix)$standard.errors
zSESFornix## (Intercept) SES
## 2 -3.462126 0.1757637
## 3 -9.318185 2.0045901pSESFornix <- (1 - pnorm(abs(zSESFornix), 0, 1)) * 2
pSESFornix## (Intercept) SES
## 2 0.0005359255 0.86047956
## 3 0.0000000000 0.04500689Trajectory
K-means Clustering
set.seed(41)
Kmeans_Shape <- kmeans(AdvDTI[,19], 2, iter.max = 10, nstart = 50)ShapeGroups <- as.factor(Kmeans_Shape$cluster)
ShapePlotCluster <- data.frame(c(AdvDTI$Fornix_FA_t1), #select FA at time 1
c(AdvDTI$Fornix_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(ShapeGroups)) #insert cluster number
colnames(ShapePlotCluster)<-c('T1', 'T2','ID','Cluster') #give column name
ShapePlotAllClusters<-melt(ShapePlotCluster,by='ID')
ggplot(ShapePlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2")))+
scale_color_manual(values = my_colors,
breaks =c("1","2"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))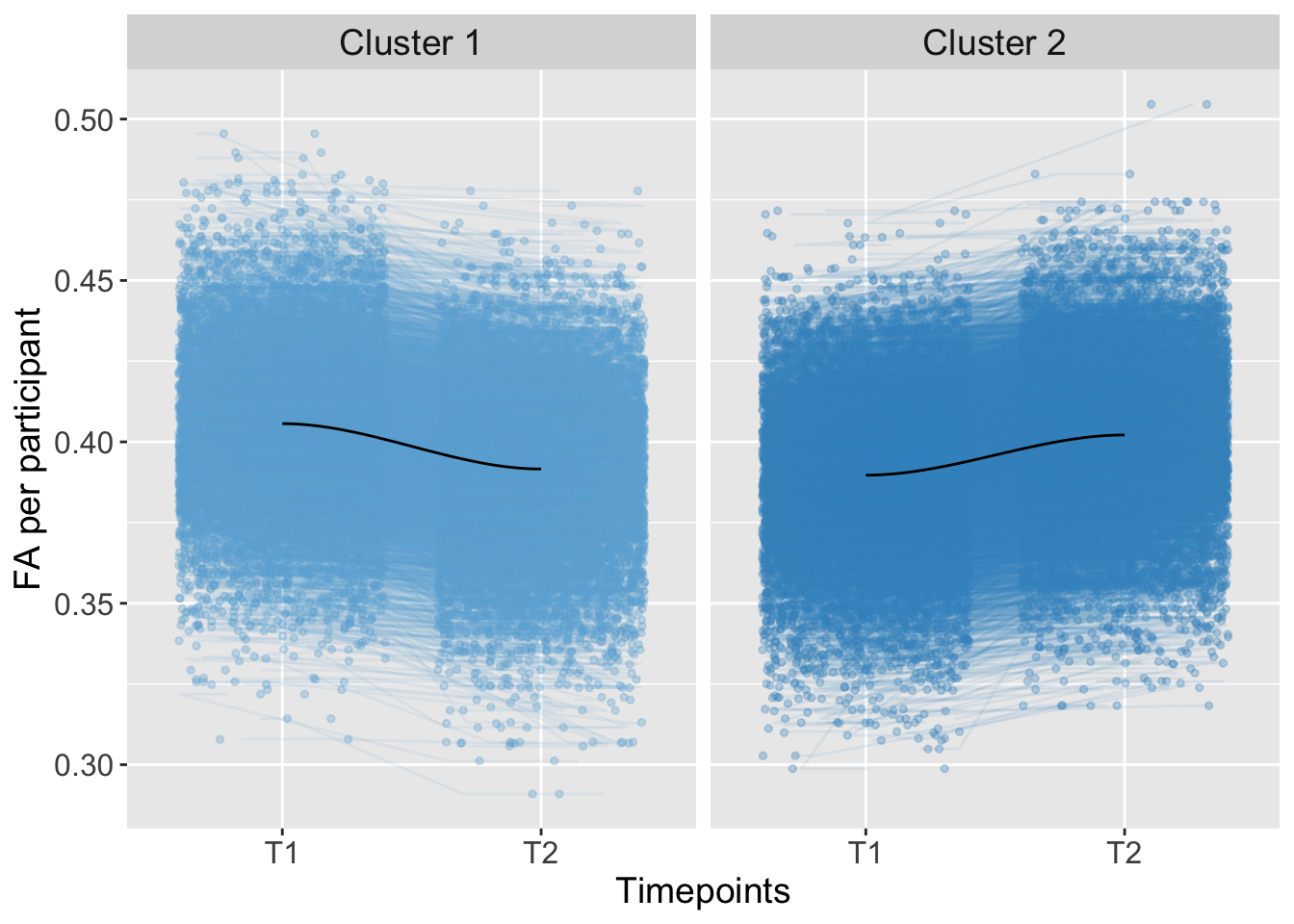
Regression results
mod_PTSD_Shape = multinom(ShapeGroups ~ PTSDSum, data=AdvDTI)
mod_School_Shape = multinom(ShapeGroups ~ SchoolSum, data=AdvDTI)
mod_Family_Shape = multinom(ShapeGroups ~ FamilySum, data=AdvDTI)
mod_Safety_Shape = multinom(ShapeGroups ~ Safety, data=AdvDTI)
mod_SES_Shape = multinom(ShapeGroups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 1.6768 1 0.1953summary(mod_PTSD_Shape)## Call:
## multinom(formula = ShapeGroups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) 0.17694746 0.01931479
## PTSDSum -0.02538289 0.01961343
##
## Residual Deviance: 14897.08
## AIC: 14901.08zPTShape <- summary(mod_PTSD_Shape)$coefficients/summary(mod_PTSD_Shape)$standard.errors
zPTShape## (Intercept) PTSDSum
## 9.161243 -1.294159pPTShape <- (1 - pnorm(abs(zPTShape), 0, 1)) * 2
pPTShape## (Intercept) PTSDSum
## 0.0000000 0.1956106Anova(mod_School_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 0.31155 1 0.5767summary(mod_School_Shape)## Call:
## multinom(formula = ShapeGroups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) 0.17705740 0.01931391
## SchoolSum 0.01086244 0.01946410
##
## Residual Deviance: 14898.44
## AIC: 14902.44zSShape <- summary(mod_School_Shape)$coefficients/summary(mod_School_Shape)$standard.errors
zSShape## (Intercept) SchoolSum
## 9.1673508 0.5580757pSShape <- (1 - pnorm(abs(zSShape), 0, 1)) * 2
pSShape## (Intercept) SchoolSum
## 0.0000000 0.5767927Anova(mod_Family_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## FamilySum 0.30405 1 0.5814summary(mod_Family_Shape)## Call:
## multinom(formula = ShapeGroups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) 0.1769231 0.01931394
## FamilySum -0.0106733 0.01935392
##
## Residual Deviance: 14898.45
## AIC: 14902.45zFShape <- summary(mod_Family_Shape)$coefficients/summary(mod_Family_Shape)$standard.errors
zFShape## (Intercept) FamilySum
## 9.1603802 -0.5514799pFShape <- (1 - pnorm(abs(zFShape), 0, 1)) * 2
pFShape## (Intercept) FamilySum
## 0.0000000 0.5813048Anova(mod_Safety_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## Safety 13.015 1 0.000309 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Safety_Shape)## Call:
## multinom(formula = ShapeGroups ~ Safety, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) 0.17690566 0.01932493
## Safety -0.07007181 0.01942252
##
## Residual Deviance: 14885.74
## AIC: 14889.74zSafShape <- summary(mod_Safety_Shape)$coefficients/summary(mod_Safety_Shape)$standard.errors
zSafShape## (Intercept) Safety
## 9.154270 -3.607761pSafShape <- (1 - pnorm(abs(zSafShape), 0, 1)) * 2
pSafShape## (Intercept) Safety
## 0.0000000000 0.0003088512Anova(mod_SES_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## SES 3.2716 1 0.07049 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_SES_Shape)## Call:
## multinom(formula = ShapeGroups ~ SES, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) 0.17664027 0.01931696
## SES -0.03518684 0.01944893
##
## Residual Deviance: 14895.48
## AIC: 14899.48zSESShape <- summary(mod_SES_Shape)$coefficients/summary(mod_SES_Shape)$standard.errors
zSESShape## (Intercept) SES
## 9.144310 -1.809192pSESShape <- (1 - pnorm(abs(zSESShape), 0, 1)) * 2
pSESShape## (Intercept) SES
## 0.00000000 0.07042124Total
K-means Clustering
set.seed(123)
Kmeans_Magnitude <- kmeans(AdvDTI[,20], 2, iter.max = 10, nstart = 50) #clusterMagnitudeGroups <- as.factor(Kmeans_Magnitude$cluster)
MagnitudePlotCluster <- data.frame(c(AdvDTI$Fornix_FA_t1), #select FA at time 1
c(AdvDTI$Fornix_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(MagnitudeGroups)) #insert cluster number
colnames(MagnitudePlotCluster)<-c('T1', 'T2','ID','Cluster') #give column name
MagnitudePlotAllClusters<-melt(MagnitudePlotCluster,by='ID')
ggplot(MagnitudePlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2")))+
scale_color_manual(values = my_colors,
breaks =c("1","2"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))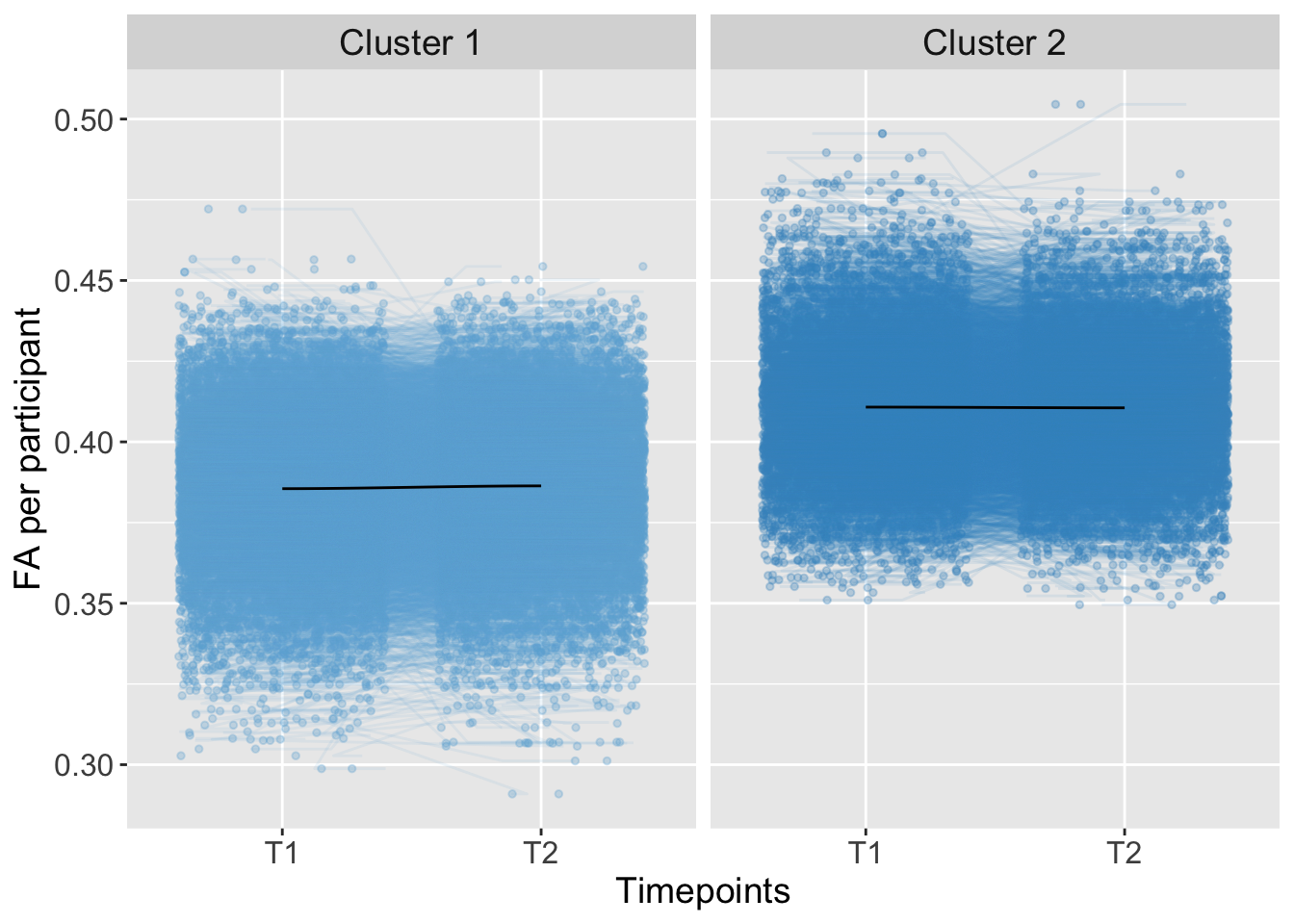
Regression results
mod_PTSD_Magnitude = multinom(MagnitudeGroups ~ PTSDSum, data=AdvDTI)
mod_School_Magnitude = multinom(MagnitudeGroups ~ SchoolSum, data=AdvDTI)
mod_Family_Magnitude = multinom(MagnitudeGroups ~ FamilySum, data=AdvDTI)
mod_Safety_Magnitude = multinom(MagnitudeGroups ~ Safety, data=AdvDTI)
mod_SES_Magnitude = multinom(MagnitudeGroups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 0.10885 1 0.7415summary(mod_PTSD_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.185972083 0.01932131
## PTSDSum -0.006484455 0.01966635
##
## Residual Deviance: 14889.93
## AIC: 14893.93zPTMagnitude <- summary(mod_PTSD_Magnitude)$coefficients/summary(mod_PTSD_Magnitude)$standard.errors
zPTMagnitude## (Intercept) PTSDSum
## -9.6252317 -0.3297233pPTMagnitude <- (1 - pnorm(abs(zPTMagnitude), 0, 1)) * 2
pPTMagnitude## (Intercept) PTSDSum
## 0.0000000 0.7416091Anova(mod_School_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 2.2083 1 0.1373summary(mod_School_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.18614024 0.01932369
## SchoolSum -0.02894984 0.01949215
##
## Residual Deviance: 14887.83
## AIC: 14891.83zSMagnitude <- summary(mod_School_Magnitude)$coefficients/summary(mod_School_Magnitude)$standard.errors
zSMagnitude## (Intercept) SchoolSum
## -9.632750 -1.485205pSMagnitude <- (1 - pnorm(abs(zSMagnitude), 0, 1)) * 2
pSMagnitude## (Intercept) SchoolSum
## 0.0000000 0.1374895Anova(mod_Family_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## FamilySum 18.945 1 0.00001345 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Family_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.18561485 0.01933809
## FamilySum 0.08422157 0.01935353
##
## Residual Deviance: 14871.1
## AIC: 14875.1zFMagnitude <- summary(mod_Family_Magnitude)$coefficients/summary(mod_Family_Magnitude)$standard.errors
zFMagnitude## (Intercept) FamilySum
## -9.598405 4.351741pFMagnitude <- (1 - pnorm(abs(zFMagnitude), 0, 1)) * 2
pFMagnitude## (Intercept) FamilySum
## 0.00000000000 0.00001350605Anova(mod_Safety_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## Safety 1.6599 1 0.1976summary(mod_Safety_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ Safety, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.18608122 0.01932298
## Safety -0.02508482 0.01948007
##
## Residual Deviance: 14888.38
## AIC: 14892.38zSafMagnitude <- summary(mod_Safety_Magnitude)$coefficients/summary(mod_Safety_Magnitude)$standard.errors
zSafMagnitude## (Intercept) Safety
## -9.630047 -1.287717pSafMagnitude <- (1 - pnorm(abs(zSafMagnitude), 0, 1)) * 2
pSafMagnitude## (Intercept) Safety
## 0.0000000 0.1978444Anova(mod_SES_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## SES 0.59363 1 0.441summary(mod_SES_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ SES, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.18613594 0.01932328
## SES -0.01501673 0.01949703
##
## Residual Deviance: 14889.45
## AIC: 14893.45zSESMagnitude <- summary(mod_SES_Magnitude)$coefficients/summary(mod_SES_Magnitude)$standard.errors
zSESMagnitude## (Intercept) SES
## -9.6327301 -0.7702061pSESMagnitude <- (1 - pnorm(abs(zSESMagnitude), 0, 1)) * 2
pSESMagnitude## (Intercept) SES
## 0.0000000 0.4411777Ayla Pollmann - 2023