All Tracts
In this script, you can find the results of the analysis based on all brain tracts. To examine individual differences, we used K-means clustering in an exploratory analysis:
K-means clustering was conducted based on the total level of brain connectivity (total; FA at imaging timepoint one plus timepoint two: higher FA total level implies greater connectivity, lower FA total levels imply lower connectivity) and the development of brain connectivity from imaging timepoint one to two (trajectory; FA at timepoint two minus timepoint one).
Multinomial logistic regression was used to predict cluster membership based on adversity experiences at baseline. To adjust for multiple comparisons, p-values were Bonferroni-corrected in the actual paper. We corrected for the number of brain regions (four) and adversity scales (five).
Total & Trajectory
K-means Clustering
set.seed(27)
Kmeans_AllTracts <- kmeans(AdvDTI[,19:20], 4, iter.max = 10, nstart = 50) Groups <- as.factor(Kmeans_AllTracts$cluster)
samplesize <- 10808
id=factor(1:samplesize)
PlotCluster <- data.frame(c(AdvDTI$AllTracts_FA_t1), #select FA at time 1
c(AdvDTI$AllTracts_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(Groups)) #insert cluster number
colnames(PlotCluster)<-c('T1','T2','ID','Cluster') #give column name
PlotAllClusters<-melt(PlotCluster,by='ID')
ggplot(PlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2","3","4")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2", "3" = "Cluster 3", "4" = "Cluster 4")))+
scale_color_manual(values = my_colors,
breaks =c("1","2","3","4"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))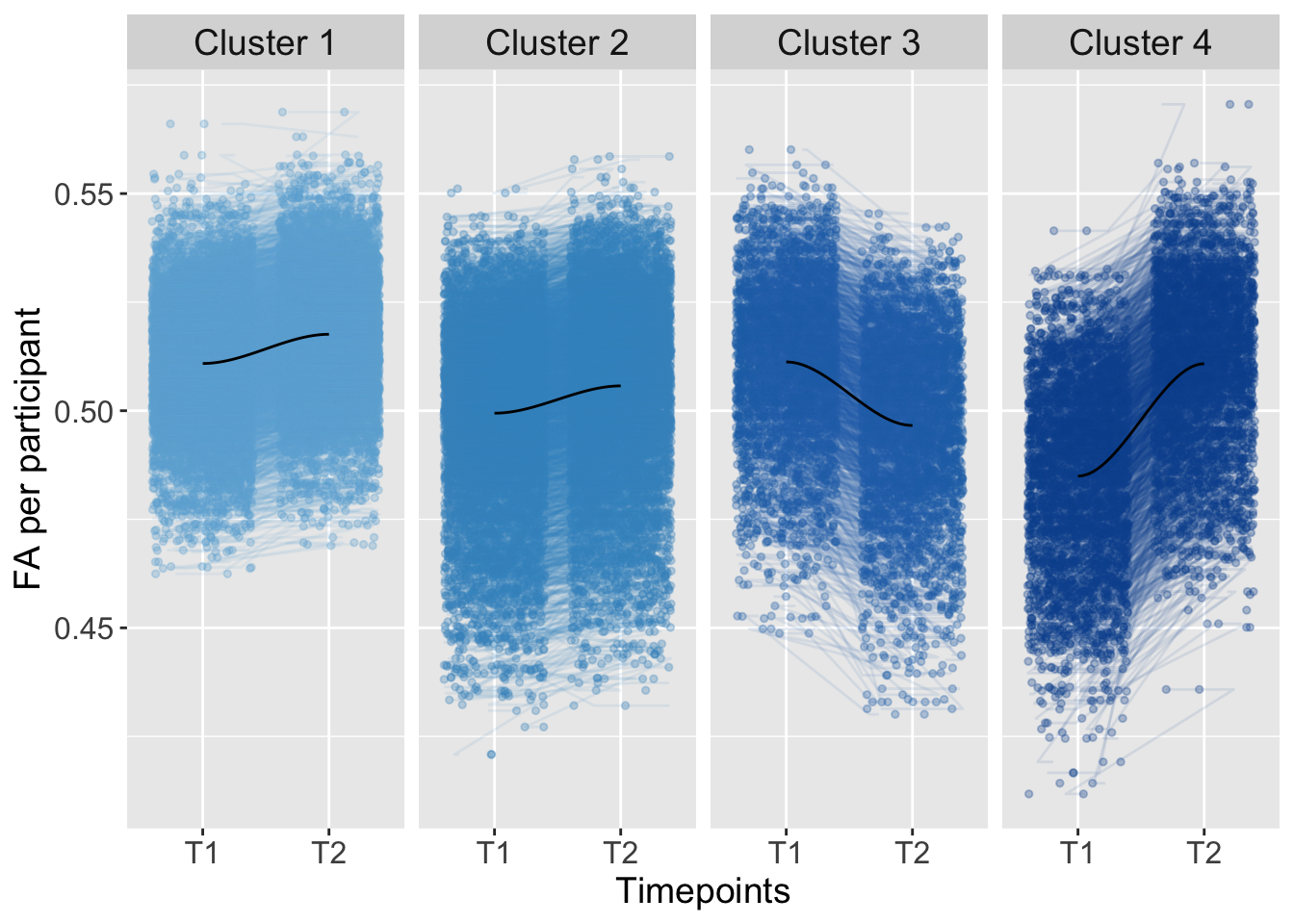
Regression results
AdvDTI$Groups <- as.factor(Kmeans_AllTracts$cluster)
AdvDTI[,2:20] <- apply(AdvDTI[,2:20],2, function(x) as.numeric(as.character(x)))
mod_PTSD_AllTracts = multinom(Groups ~ PTSDSum, data=AdvDTI)
mod_School_AllTracts = multinom(Groups ~ SchoolSum, data=AdvDTI)
mod_Family_AllTracts = multinom(Groups ~ FamilySum, data=AdvDTI)
mod_Safety_AllTracts = multinom(Groups ~ Safety, data=AdvDTI)
mod_SES_AllTracts = multinom(Groups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_AllTracts, type=3) ## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 2.8304 3 0.4185summary(mod_PTSD_AllTracts)## Call:
## multinom(formula = Groups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) PTSDSum
## 2 -0.005822554 -0.01273717
## 3 -0.937476354 0.03236716
## 4 -0.950441146 0.02157508
##
## Std. Errors:
## (Intercept) PTSDSum
## 2 0.02268577 0.02364157
## 3 0.03019692 0.02933320
## 4 0.03033596 0.03003556
##
## Residual Deviance: 27811.95
## AIC: 27823.95zPTAllTracts <- summary(mod_PTSD_AllTracts)$coefficients/summary(mod_PTSD_AllTracts)$standard.errors
zPTAllTracts## (Intercept) PTSDSum
## 2 -0.2566611 -0.5387618
## 3 -31.0454345 1.1034310
## 4 -31.3305161 0.7183180pPTAllTracts <- (1 - pnorm(abs(zPTAllTracts), 0, 1)) * 2
pPTAllTracts## (Intercept) PTSDSum
## 2 0.7974404 0.5900512
## 3 0.0000000 0.2698400
## 4 0.0000000 0.4725612Anova(mod_School_AllTracts, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 0.17147 3 0.9821summary(mod_School_AllTracts)## Call:
## multinom(formula = Groups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) SchoolSum
## 2 -0.005612845 0.009078191
## 3 -0.937148039 0.001383108
## 4 -0.950300335 0.004986463
##
## Std. Errors:
## (Intercept) SchoolSum
## 2 0.02268396 0.02284740
## 3 0.03019084 0.03045130
## 4 0.03033386 0.03056388
##
## Residual Deviance: 27814.61
## AIC: 27826.61zSAllTracts <- summary(mod_School_AllTracts)$coefficients/summary(mod_School_AllTracts)$standard.errors
zSAllTracts## (Intercept) SchoolSum
## 2 -0.2474367 0.39734019
## 3 -31.0408049 0.04542034
## 4 -31.3280402 0.16314889pSAllTracts <- (1 - pnorm(abs(zSAllTracts), 0, 1)) * 2
pSAllTracts## (Intercept) SchoolSum
## 2 0.8045703 0.6911166
## 3 0.0000000 0.9637723
## 4 0.0000000 0.8704012Anova(mod_Family_AllTracts, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## FamilySum 14.828 3 0.00197 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Family_AllTracts)## Call:
## multinom(formula = Groups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## (Intercept) FamilySum
## 2 -0.006146027 -0.08666201
## 3 -0.936660296 -0.05799022
## 4 -0.949658294 -0.04421635
##
## Std. Errors:
## (Intercept) FamilySum
## 2 0.02270519 0.02277359
## 3 0.03020029 0.03024979
## 4 0.03033979 0.03024833
##
## Residual Deviance: 27799.96
## AIC: 27811.96zFAllTracts <- summary(mod_Family_AllTracts)$coefficients/summary(mod_Family_AllTracts)$standard.errors
zFAllTracts## (Intercept) FamilySum
## 2 -0.2706882 -3.805373
## 3 -31.0149419 -1.917045
## 4 -31.3007518 -1.461778pFAllTracts <- (1 - pnorm(abs(zFAllTracts), 0, 1)) * 2
pFAllTracts## (Intercept) FamilySum
## 2 0.7866309 0.0001415906
## 3 0.0000000 0.0552321820
## 4 0.0000000 0.1438019528Anova(mod_Safety_AllTracts, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## Safety 5.1884 3 0.1585summary(mod_Safety_AllTracts)## Call:
## multinom(formula = Groups ~ Safety, data = AdvDTI)
##
## Coefficients:
## (Intercept) Safety
## 2 -0.006025944 -0.045886379
## 3 -0.937280943 0.006819441
## 4 -0.950283841 -0.024853312
##
## Std. Errors:
## (Intercept) Safety
## 2 0.02269015 0.02290123
## 3 0.03019564 0.03004247
## 4 0.03033536 0.03053461
##
## Residual Deviance: 27809.6
## AIC: 27821.6zSafAllTracts <- summary(mod_Safety_AllTracts)$coefficients/summary(mod_Safety_AllTracts)$standard.errors
zSafAllTracts## (Intercept) Safety
## 2 -0.2655753 -2.0036644
## 3 -31.0402713 0.2269934
## 4 -31.3259420 -0.8139390pSafAllTracts <- (1 - pnorm(abs(zSafAllTracts), 0, 1)) * 2
pSafAllTracts## (Intercept) Safety
## 2 0.7905663 0.04510603
## 3 0.0000000 0.82042887
## 4 0.0000000 0.41567989Anova(mod_SES_AllTracts, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## SES 24.309 3 0.00002153 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_SES_AllTracts)## Call:
## multinom(formula = Groups ~ SES, data = AdvDTI)
##
## Coefficients:
## (Intercept) SES
## 2 -0.007124469 -0.03855303
## 3 -0.940341713 0.10095172
## 4 -0.950492577 0.04607389
##
## Std. Errors:
## (Intercept) SES
## 2 0.02270501 0.02328433
## 3 0.03026231 0.02938147
## 4 0.03034247 0.03018164
##
## Residual Deviance: 27790.48
## AIC: 27802.48zSESAllTracts <- summary(mod_SES_AllTracts)$coefficients/summary(mod_SES_AllTracts)$standard.errors
zSESAllTracts## (Intercept) SES
## 2 -0.313784 -1.655750
## 3 -31.073027 3.435897
## 4 -31.325480 1.526554pSESAllTracts <- (1 - pnorm(abs(zSESAllTracts), 0, 1)) * 2
pSESAllTracts## (Intercept) SES
## 2 0.7536851 0.0977724318
## 3 0.0000000 0.0005905949
## 4 0.0000000 0.1268720092Trajectory
K-means Clustering
set.seed(4)
Kmeans_Shape <- kmeans(AdvDTI[,19], 3, iter.max = 10, nstart = 50) ShapeGroups <- as.factor(Kmeans_Shape$cluster)
ShapePlotCluster <- data.frame(c(AdvDTI$AllTracts_FA_t1), #select FA at time 1
c(AdvDTI$AllTracts_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(ShapeGroups)) #insert cluster number
colnames(ShapePlotCluster)<-c('T1', 'T2','ID','Cluster') #give column name
ShapePlotAllClusters<-melt(ShapePlotCluster,by='ID')
ggplot(ShapePlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2","3")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2", "3" = "Cluster 3")))+
scale_color_manual(values = my_colors,
breaks =c("1","2","3"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))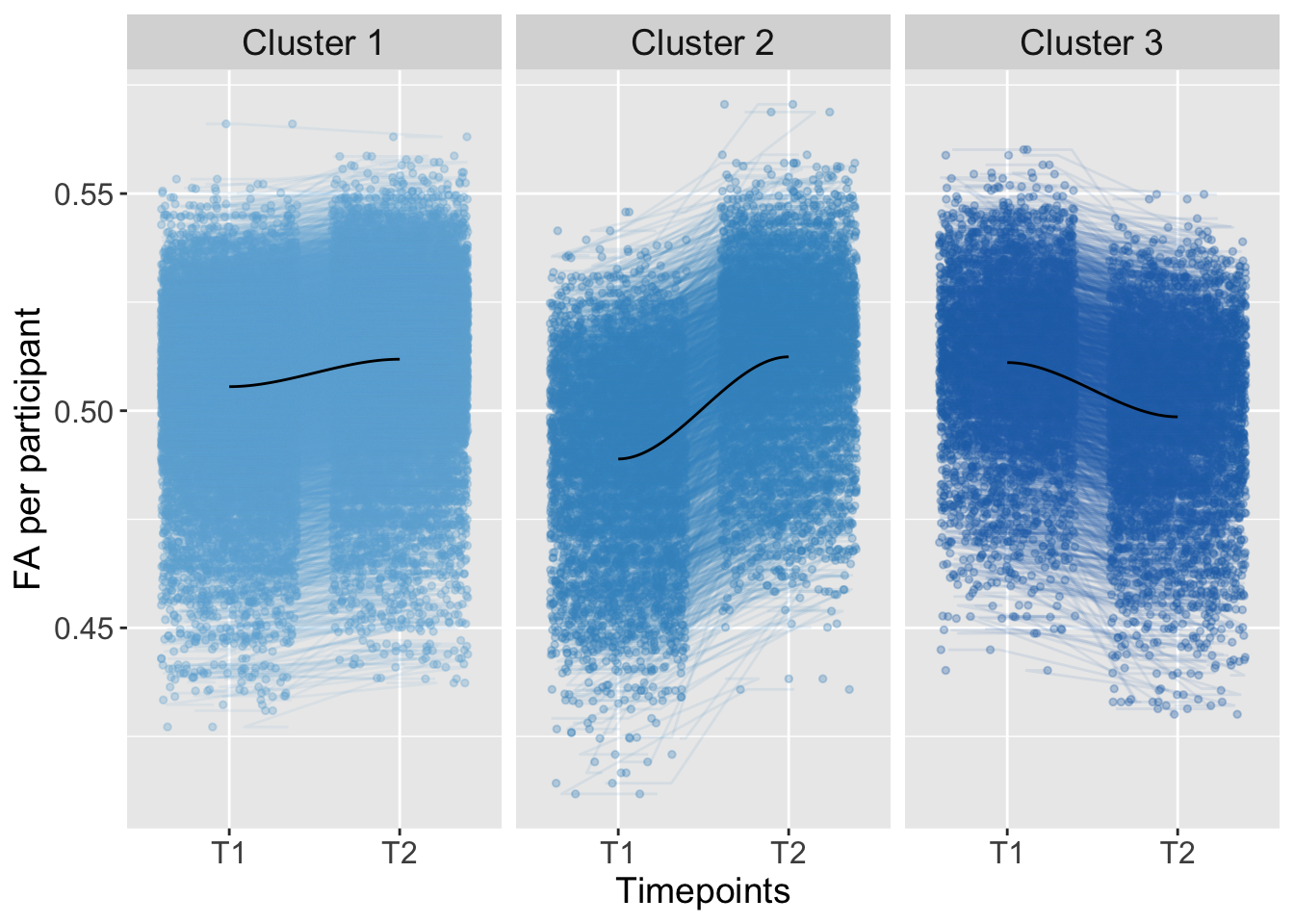
Regression results
mod_PTSD_Shape = multinom(ShapeGroups ~ PTSDSum, data=AdvDTI)
mod_School_Shape = multinom(ShapeGroups ~ SchoolSum, data=AdvDTI)
mod_Family_Shape = multinom(ShapeGroups ~ FamilySum, data=AdvDTI)
mod_Safety_Shape = multinom(ShapeGroups ~ Safety, data=AdvDTI)
mod_SES_Shape = multinom(ShapeGroups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 5.067 2 0.07938 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_PTSD_Shape)## Call:
## multinom(formula = ShapeGroups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) PTSDSum
## 2 -1.085692 0.0161057
## 3 -1.148634 0.0554215
##
## Std. Errors:
## (Intercept) PTSDSum
## 2 0.02463092 0.02532587
## 3 0.02522796 0.02424196
##
## Residual Deviance: 20429.51
## AIC: 20437.51zPTShape <- summary(mod_PTSD_Shape)$coefficients/summary(mod_PTSD_Shape)$standard.errors
zPTShape## (Intercept) PTSDSum
## 2 -44.07842 0.6359386
## 3 -45.53018 2.2861810pPTShape <- (1 - pnorm(abs(zPTShape), 0, 1)) * 2
pPTShape## (Intercept) PTSDSum
## 2 0 0.52481644
## 3 0 0.02224367Anova(mod_School_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 0.059123 2 0.9709summary(mod_School_Shape)## Call:
## multinom(formula = ShapeGroups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) SchoolSum
## 2 -1.085773 0.003553326
## 3 -1.148053 0.005733582
##
## Std. Errors:
## (Intercept) SchoolSum
## 2 0.02462978 0.02480264
## 3 0.02521440 0.02537409
##
## Residual Deviance: 20434.52
## AIC: 20442.52zSShape <- summary(mod_School_Shape)$coefficients/summary(mod_School_Shape)$standard.errors
zSShape## (Intercept) SchoolSum
## 2 -44.08373 0.143264
## 3 -45.53163 0.225962pSShape <- (1 - pnorm(abs(zSShape), 0, 1)) * 2
pSShape## (Intercept) SchoolSum
## 2 0 0.8860817
## 3 0 0.8212309Anova(mod_Family_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## FamilySum 0.89571 2 0.639summary(mod_Family_Shape)## Call:
## multinom(formula = ShapeGroups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## (Intercept) FamilySum
## 2 -1.085746 0.01364330
## 3 -1.147974 0.02220501
##
## Std. Errors:
## (Intercept) FamilySum
## 2 0.02463064 0.02465685
## 3 0.02521510 0.02516377
##
## Residual Deviance: 20433.68
## AIC: 20441.68zFShape <- summary(mod_Family_Shape)$coefficients/summary(mod_Family_Shape)$standard.errors
zFShape## (Intercept) FamilySum
## 2 -44.08112 0.5533272
## 3 -45.52726 0.8824198pFShape <- (1 - pnorm(abs(zFShape), 0, 1)) * 2
pFShape## (Intercept) FamilySum
## 2 0 0.5800394
## 3 0 0.3775498Anova(mod_Safety_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## Safety 1.9329 2 0.3804summary(mod_Safety_Shape)## Call:
## multinom(formula = ShapeGroups ~ Safety, data = AdvDTI)
##
## Coefficients:
## (Intercept) Safety
## 2 -1.085924 -0.006641791
## 3 -1.148280 0.031650394
##
## Std. Errors:
## (Intercept) Safety
## 2 0.02463234 0.02492175
## 3 0.02521915 0.02514743
##
## Residual Deviance: 20432.64
## AIC: 20440.64zSafShape <- summary(mod_Safety_Shape)$coefficients/summary(mod_Safety_Shape)$standard.errors
zSafShape## (Intercept) Safety
## 2 -44.08530 -0.2665058
## 3 -45.53206 1.2585935pSafShape <- (1 - pnorm(abs(zSafShape), 0, 1)) * 2
pSafShape## (Intercept) Safety
## 2 0 0.7898497
## 3 0 0.2081772Anova(mod_SES_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## SES 26.062 2 0.000002191 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_SES_Shape)## Call:
## multinom(formula = ShapeGroups ~ SES, data = AdvDTI)
##
## Coefficients:
## (Intercept) SES
## 2 -1.084748 0.05788339
## 3 -1.149835 0.12432410
##
## Std. Errors:
## (Intercept) SES
## 2 0.02463816 0.02479614
## 3 0.02527547 0.02469067
##
## Residual Deviance: 20408.51
## AIC: 20416.51zSESShape <- summary(mod_SES_Shape)$coefficients/summary(mod_SES_Shape)$standard.errors
zSESShape## (Intercept) SES
## 2 -44.02713 2.334371
## 3 -45.49211 5.035265pSESShape <- (1 - pnorm(abs(zSESShape), 0, 1)) * 2
pSESShape## (Intercept) SES
## 2 0 0.0195763167781
## 3 0 0.0000004771871Total
K-means Clustering
set.seed(123)
Kmeans_Magnitude <- kmeans(AdvDTI[,20], 2, iter.max = 10, nstart = 50) #clusterMagnitudeGroups <- as.factor(Kmeans_Magnitude$cluster)
MagnitudePlotCluster <- data.frame(c(AdvDTI$AllTracts_FA_t1), #select FA at time 1
c(AdvDTI$AllTracts_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(MagnitudeGroups)) #insert cluster number
colnames(MagnitudePlotCluster)<-c('T1', 'T2','ID','Cluster') #give column name
MagnitudePlotAllClusters<-melt(MagnitudePlotCluster,by='ID')
ggplot(MagnitudePlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2")), labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2")))+
scale_color_manual(values = my_colors,
breaks =c("1","2"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))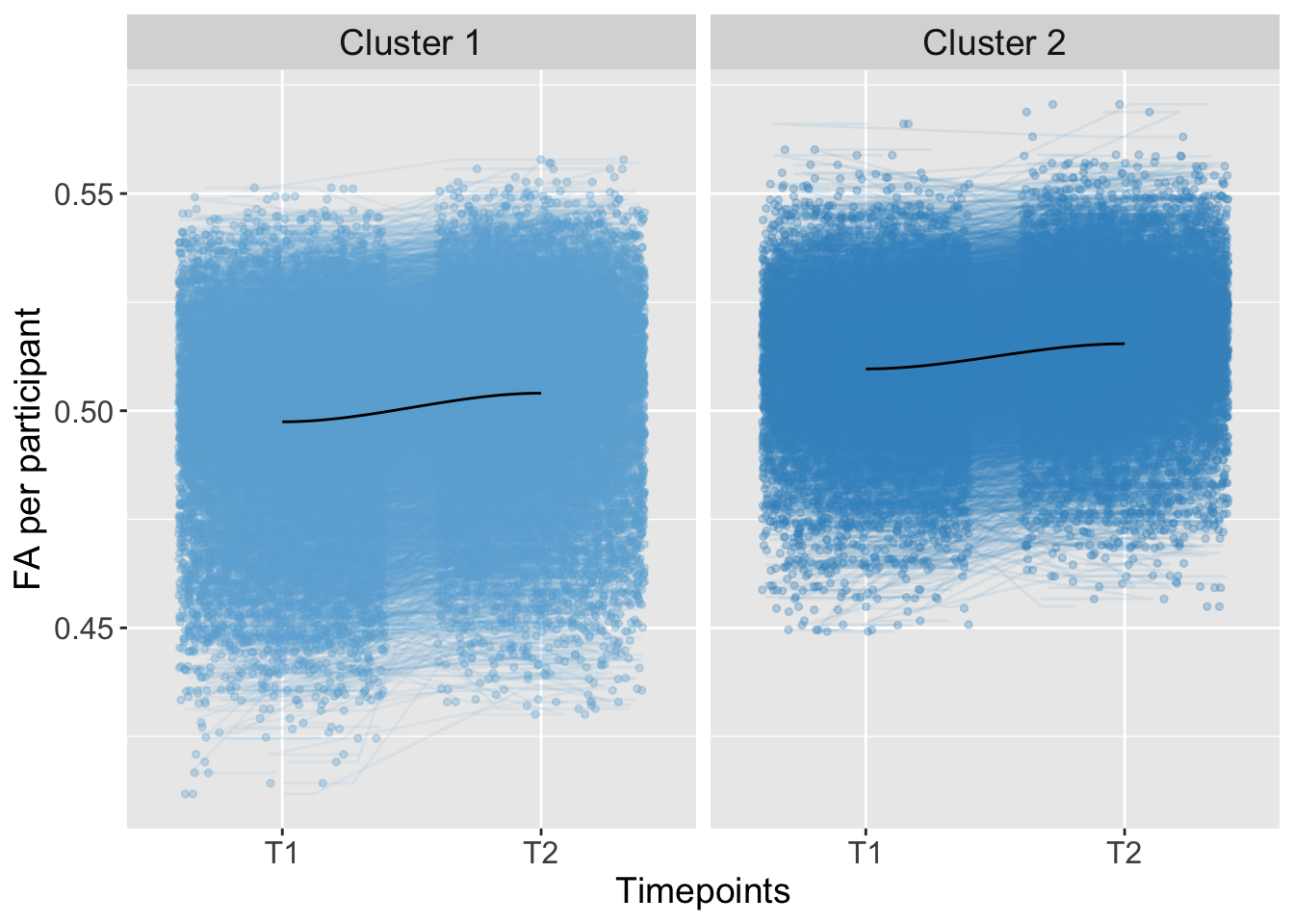
Regression results
mod_PTSD_Magnitude = multinom(MagnitudeGroups ~ PTSDSum, data=AdvDTI)
mod_School_Magnitude = multinom(MagnitudeGroups ~ SchoolSum, data=AdvDTI)
mod_Family_Magnitude = multinom(MagnitudeGroups ~ FamilySum, data=AdvDTI)
mod_Safety_Magnitude = multinom(MagnitudeGroups ~ Safety, data=AdvDTI)
mod_SES_Magnitude = multinom(MagnitudeGroups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 0.0017071 1 0.967summary(mod_PTSD_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.1074295514 0.01926572
## PTSDSum 0.0008087193 0.01957425
##
## Residual Deviance: 14951.93
## AIC: 14955.93zPTMagnitude <- summary(mod_PTSD_Magnitude)$coefficients/summary(mod_PTSD_Magnitude)$standard.errors
zPTMagnitude## (Intercept) PTSDSum
## -5.57620161 0.04131546pPTMagnitude <- (1 - pnorm(abs(zPTMagnitude), 0, 1)) * 2
pPTMagnitude## (Intercept) PTSDSum
## 0.0000000245827 0.9670444055551Anova(mod_School_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 1.0618 1 0.3028summary(mod_School_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.10754473 0.01926698
## SchoolSum -0.02000606 0.01941959
##
## Residual Deviance: 14950.87
## AIC: 14954.87zSMagnitude <- summary(mod_School_Magnitude)$coefficients/summary(mod_School_Magnitude)$standard.errors
zSMagnitude## (Intercept) SchoolSum
## -5.581817 -1.030200pSMagnitude <- (1 - pnorm(abs(zSMagnitude), 0, 1)) * 2
pSMagnitude## (Intercept) SchoolSum
## 0.00000002380189 0.30291636150292Anova(mod_Family_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## FamilySum 16.813 1 0.00004125 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Family_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.10697844 0.01928086
## FamilySum 0.07918894 0.01932407
##
## Residual Deviance: 14935.12
## AIC: 14939.12zFMagnitude <- summary(mod_Family_Magnitude)$coefficients/summary(mod_Family_Magnitude)$standard.errors
zFMagnitude## (Intercept) FamilySum
## -5.548427 4.097944pFMagnitude <- (1 - pnorm(abs(zFMagnitude), 0, 1)) * 2
pFMagnitude## (Intercept) FamilySum
## 0.00000002882511 0.00004168363914Anova(mod_Safety_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## Safety 3.5816 1 0.05842 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Safety_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ Safety, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.10731769 0.01926889
## Safety 0.03669357 0.01938778
##
## Residual Deviance: 14948.35
## AIC: 14952.35zSafMagnitude <- summary(mod_Safety_Magnitude)$coefficients/summary(mod_Safety_Magnitude)$standard.errors
zSafMagnitude## (Intercept) Safety
## -5.569479 1.892613pSafMagnitude <- (1 - pnorm(abs(zSafMagnitude), 0, 1)) * 2
pSafMagnitude## (Intercept) Safety
## 0.00000002555026 0.05840932338287Anova(mod_SES_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## SES 0.19948 1 0.6551summary(mod_SES_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ SES, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.107333027 0.01926703
## SES 0.008673255 0.01941782
##
## Residual Deviance: 14951.73
## AIC: 14955.73zSESMagnitude <- summary(mod_SES_Magnitude)$coefficients/summary(mod_SES_Magnitude)$standard.errors
zSESMagnitude## (Intercept) SES
## -5.5708138 0.4466646pSESMagnitude <- (1 - pnorm(abs(zSESMagnitude), 0, 1)) * 2
pSESMagnitude## (Intercept) SES
## 0.00000002535522 0.65511722427027Ayla Pollmann - 2023