ATR
In this script, you can find the results of the analysis across the ATR. To examine individual differences, we used K-means clustering in an exploratory analysis:
K-means clustering was conducted based on the total level of brain connectivity (total; FA at imaging timepoint one plus timepoint two: higher FA total level implies greater connectivity, lower FA total levels imply lower connectivity) and the development of brain connectivity from imaging timepoint one to two (trajectory; FA at timepoint two minus timepoint one).
Multinomial logistic regression was used to predict cluster membership based on adversity experiences at baseline. To adjust for multiple comparisons, p-values were Bonferroni-corrected in the actual paper. We corrected for the number of brain regions (four) and adversity scales (five).
Total & Trajectory
K-means Clustering
set.seed(48)
Kmeans_ATR <- kmeans(AdvDTI[,19:20], 3, iter.max = 10, nstart = 50) Groups <- as.factor(Kmeans_ATR$cluster)
samplesize <- 10808
id=factor(1:samplesize)
PlotCluster <- data.frame(c(AdvDTI$ATR_FA_t1), #select FA at time 1
c(AdvDTI$ATR_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(Groups)) #insert cluster number
colnames(PlotCluster)<-c('T1','T2','ID','Cluster') #give column name
PlotAllClusters<-melt(PlotCluster,by='ID')
ggplot(PlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2","3")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2", "3" = "Cluster 3")))+
scale_color_manual(values = my_colors,
breaks =c("1","2","3"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))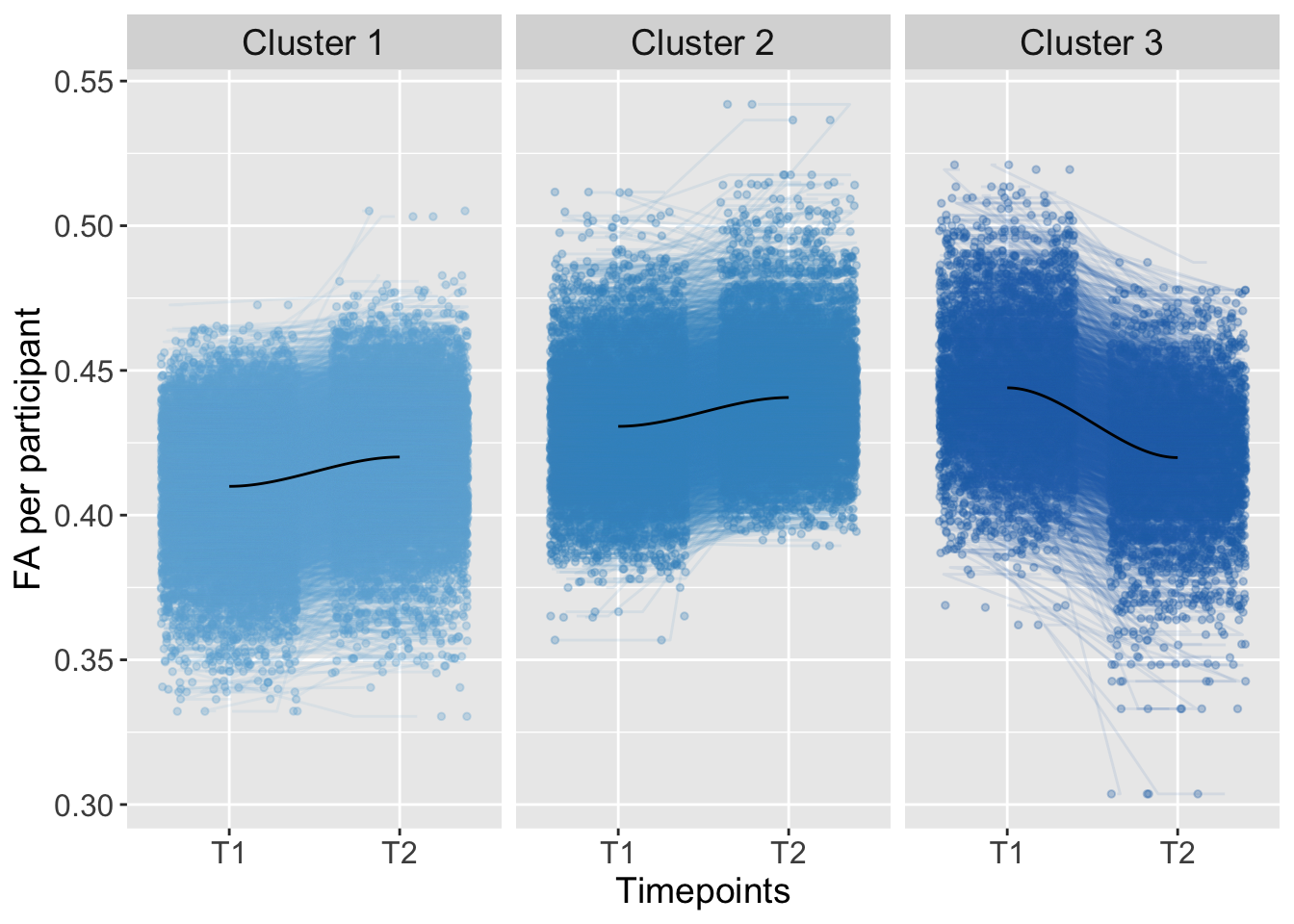
Regression results
AdvDTI$Groups <- as.factor(Kmeans_ATR$cluster)
AdvDTI[,2:20] <- apply(AdvDTI[,2:20],2, function(x) as.numeric(as.character(x)))
mod_PTSD_ATR = multinom(Groups ~ PTSDSum, data=AdvDTI)
mod_School_ATR = multinom(Groups ~ SchoolSum, data=AdvDTI)
mod_Family_ATR = multinom(Groups ~ FamilySum, data=AdvDTI)
mod_Safety_ATR = multinom(Groups ~ Safety, data=AdvDTI)
mod_SES_ATR = multinom(Groups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_ATR, type=3) ## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 4.2394 2 0.1201summary(mod_PTSD_ATR)## Call:
## multinom(formula = Groups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) PTSDSum
## 2 -0.2146444 -0.01971743
## 3 -0.7559923 0.03490151
##
## Std. Errors:
## (Intercept) PTSDSum
## 2 0.02172175 0.02277069
## 3 0.02568091 0.02482291
##
## Residual Deviance: 22793.05
## AIC: 22801.05zPTATR <- summary(mod_PTSD_ATR)$coefficients/summary(mod_PTSD_ATR)$standard.errors
zPTATR## (Intercept) PTSDSum
## 2 -9.881542 -0.8659127
## 3 -29.437913 1.4060202pPTATR <- (1 - pnorm(abs(zPTATR), 0, 1)) * 2
pPTATR## (Intercept) PTSDSum
## 2 0 0.3865381
## 3 0 0.1597181Anova(mod_School_ATR, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 0.49359 2 0.7813summary(mod_School_ATR)## Call:
## multinom(formula = Groups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) SchoolSum
## 2 -0.2143976 -0.004425915
## 3 -0.7556755 -0.018123185
##
## Std. Errors:
## (Intercept) SchoolSum
## 2 0.02171848 0.02185373
## 3 0.02567557 0.02593779
##
## Residual Deviance: 22796.8
## AIC: 22804.8zSATR <- summary(mod_School_ATR)$coefficients/summary(mod_School_ATR)$standard.errors
zSATR## (Intercept) SchoolSum
## 2 -9.871667 -0.2025244
## 3 -29.431689 -0.6987175pSATR <- (1 - pnorm(abs(zSATR), 0, 1)) * 2
pSATR## (Intercept) SchoolSum
## 2 0 0.8395068
## 3 0 0.4847286Anova(mod_Family_ATR, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## FamilySum 22.615 2 0.00001228 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Family_ATR)## Call:
## multinom(formula = Groups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## (Intercept) FamilySum
## 2 -0.2141819 0.10329844
## 3 -0.7541244 0.04402686
##
## Std. Errors:
## (Intercept) FamilySum
## 2 0.02174713 0.02173409
## 3 0.02568563 0.02596928
##
## Residual Deviance: 22774.68
## AIC: 22782.68zFATR <- summary(mod_Family_ATR)$coefficients/summary(mod_Family_ATR)$standard.errors
zFATR## (Intercept) FamilySum
## 2 -9.848742 4.752830
## 3 -29.359785 1.695344pFATR <- (1 - pnorm(abs(zFATR), 0, 1)) * 2
pFATR## (Intercept) FamilySum
## 2 0 0.000002005887
## 3 0 0.090010258416Anova(mod_Safety_ATR, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## Safety 12.226 2 0.002214 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Safety_ATR)## Call:
## multinom(formula = Groups ~ Safety, data = AdvDTI)
##
## Coefficients:
## (Intercept) Safety
## 2 -0.2139974 0.01999168
## 3 -0.7568090 0.08861226
##
## Std. Errors:
## (Intercept) Safety
## 2 0.02172327 0.02204129
## 3 0.02570615 0.02545498
##
## Residual Deviance: 22785.06
## AIC: 22793.06zSafATR <- summary(mod_Safety_ATR)$coefficients/summary(mod_Safety_ATR)$standard.errors
zSafATR## (Intercept) Safety
## 2 -9.851069 0.9070102
## 3 -29.440779 3.4811362pSafATR <- (1 - pnorm(abs(zSafATR), 0, 1)) * 2
pSafATR## (Intercept) Safety
## 2 0 0.3644013919
## 3 0 0.0004992915Anova(mod_SES_ATR, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## SES 43.98 2 0.0000000002817 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_SES_ATR)## Call:
## multinom(formula = Groups ~ SES, data = AdvDTI)
##
## Coefficients:
## (Intercept) SES
## 2 -0.2136197 0.01773418
## 3 -0.7600869 0.16094916
##
## Std. Errors:
## (Intercept) SES
## 2 0.02173987 0.02237098
## 3 0.02578426 0.02508210
##
## Residual Deviance: 22753.31
## AIC: 22761.31zSESATR <- summary(mod_SES_ATR)$coefficients/summary(mod_SES_ATR)$standard.errors
zSESATR## (Intercept) SES
## 2 -9.826171 0.7927318
## 3 -29.478715 6.4168922pSESATR <- (1 - pnorm(abs(zSESATR), 0, 1)) * 2
pSESATR## (Intercept) SES
## 2 0 0.4279340936262708
## 3 0 0.0000000001390843Trajectory
K-means Clustering
set.seed(413)
Kmeans_Shape <- kmeans(AdvDTI[,19], 2, iter.max = 10, nstart = 50)ShapeGroups <- as.factor(Kmeans_Shape$cluster)
ShapePlotCluster <- data.frame(c(AdvDTI$ATR_FA_t1), #select FA at time 1
c(AdvDTI$ATR_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(ShapeGroups)) #insert cluster number
colnames(ShapePlotCluster)<-c('T1', 'T2','ID','Cluster') #give column name
ShapePlotAllClusters<-melt(ShapePlotCluster,by='ID')
ggplot(ShapePlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2","3")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2", "3" = "Cluster 3")))+
scale_color_manual(values = my_colors,
breaks =c("1","2","3"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))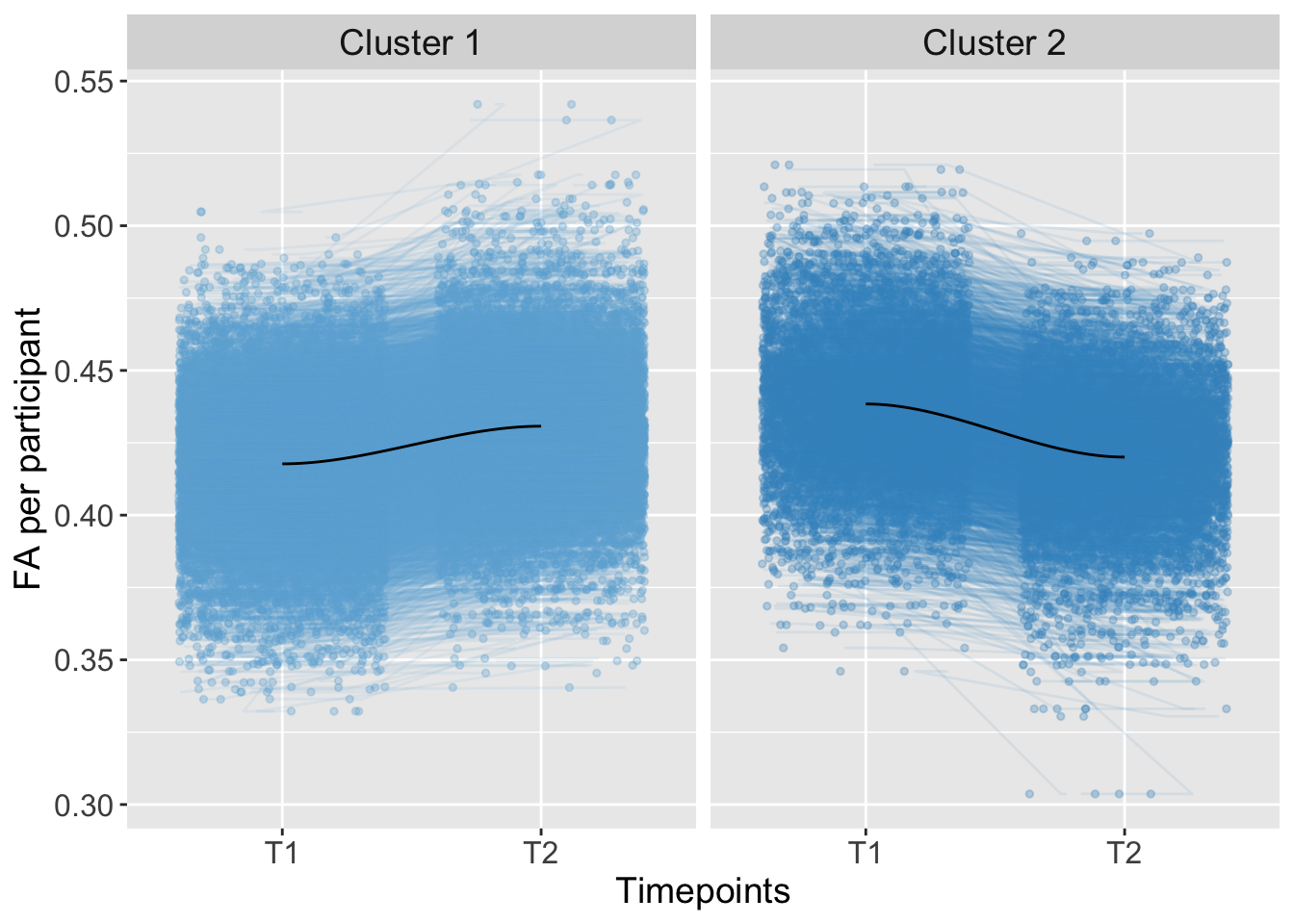
Regression results
mod_PTSD_Shape = multinom(ShapeGroups ~ PTSDSum, data=AdvDTI)
mod_School_Shape = multinom(ShapeGroups ~ SchoolSum, data=AdvDTI)
mod_Family_Shape = multinom(ShapeGroups ~ FamilySum, data=AdvDTI)
mod_Safety_Shape = multinom(ShapeGroups ~ Safety, data=AdvDTI)
mod_SES_Shape = multinom(ShapeGroups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 0.36306 1 0.5468summary(mod_PTSD_Shape)## Call:
## multinom(formula = ShapeGroups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.75910793 0.02064072
## PTSDSum 0.01255256 0.02076735
##
## Residual Deviance: 13531.05
## AIC: 13535.05zPTShape <- summary(mod_PTSD_Shape)$coefficients/summary(mod_PTSD_Shape)$standard.errors
zPTShape## (Intercept) PTSDSum
## -36.7772036 0.6044371pPTShape <- (1 - pnorm(abs(zPTShape), 0, 1)) * 2
pPTShape## (Intercept) PTSDSum
## 0.000000 0.545553Anova(mod_School_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 0.000016227 1 0.9968summary(mod_School_Shape)## Call:
## multinom(formula = ShapeGroups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.75911515422 0.02064062
## SchoolSum 0.00005720589 0.02079214
##
## Residual Deviance: 13531.41
## AIC: 13535.41zSShape <- summary(mod_School_Shape)$coefficients/summary(mod_School_Shape)$standard.errors
zSShape## (Intercept) SchoolSum
## -36.777733252 0.002751323pSShape <- (1 - pnorm(abs(zSShape), 0, 1)) * 2
pSShape## (Intercept) SchoolSum
## 0.0000000 0.9978048Anova(mod_Family_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## FamilySum 0.59641 1 0.4399summary(mod_Family_Shape)## Call:
## multinom(formula = ShapeGroups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.75928118 0.02064272
## FamilySum -0.01600978 0.02075080
##
## Residual Deviance: 13530.81
## AIC: 13534.81zFShape <- summary(mod_Family_Shape)$coefficients/summary(mod_Family_Shape)$standard.errors
zFShape## (Intercept) FamilySum
## -36.782023 -0.771526pFShape <- (1 - pnorm(abs(zFShape), 0, 1)) * 2
pFShape## (Intercept) FamilySum
## 0.0000000 0.4403952Anova(mod_Safety_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## Safety 2.8418 1 0.09184 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Safety_Shape)## Call:
## multinom(formula = ShapeGroups ~ Safety, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.75919432 0.02064379
## Safety 0.03488889 0.02065784
##
## Residual Deviance: 13528.57
## AIC: 13532.57zSafShape <- summary(mod_Safety_Shape)$coefficients/summary(mod_Safety_Shape)$standard.errors
zSafShape## (Intercept) Safety
## -36.775912 1.688893pSafShape <- (1 - pnorm(abs(zSafShape), 0, 1)) * 2
pSafShape## (Intercept) Safety
## 0.00000000 0.09123985Anova(mod_SES_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## SES 32.551 1 0.00000001161 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_SES_Shape)## Call:
## multinom(formula = ShapeGroups ~ SES, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.7601029 0.02067952
## SES 0.1171676 0.02044286
##
## Residual Deviance: 13498.86
## AIC: 13502.86zSESShape <- summary(mod_SES_Shape)$coefficients/summary(mod_SES_Shape)$standard.errors
zSESShape## (Intercept) SES
## -36.756318 5.731468pSESShape <- (1 - pnorm(abs(zSESShape), 0, 1)) * 2
pSESShape## (Intercept) SES
## 0.000000000000000 0.000000009956537Total
K-means Clustering
set.seed(123)
Kmeans_Magnitude <- kmeans(AdvDTI[,20], 2, iter.max = 10, nstart = 50) #clusterMagnitudeGroups <- as.factor(Kmeans_Magnitude$cluster)
MagnitudePlotCluster <- data.frame(c(AdvDTI$ATR_FA_t1), #select FA at time 1
c(AdvDTI$ATR_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(MagnitudeGroups)) #insert cluster number
colnames(MagnitudePlotCluster)<-c('T1', 'T2','ID','Cluster') #give column name
MagnitudePlotAllClusters<-melt(MagnitudePlotCluster,by='ID')
ggplot(MagnitudePlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2")))+
scale_color_manual(values = my_colors,
breaks =c("1","2"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))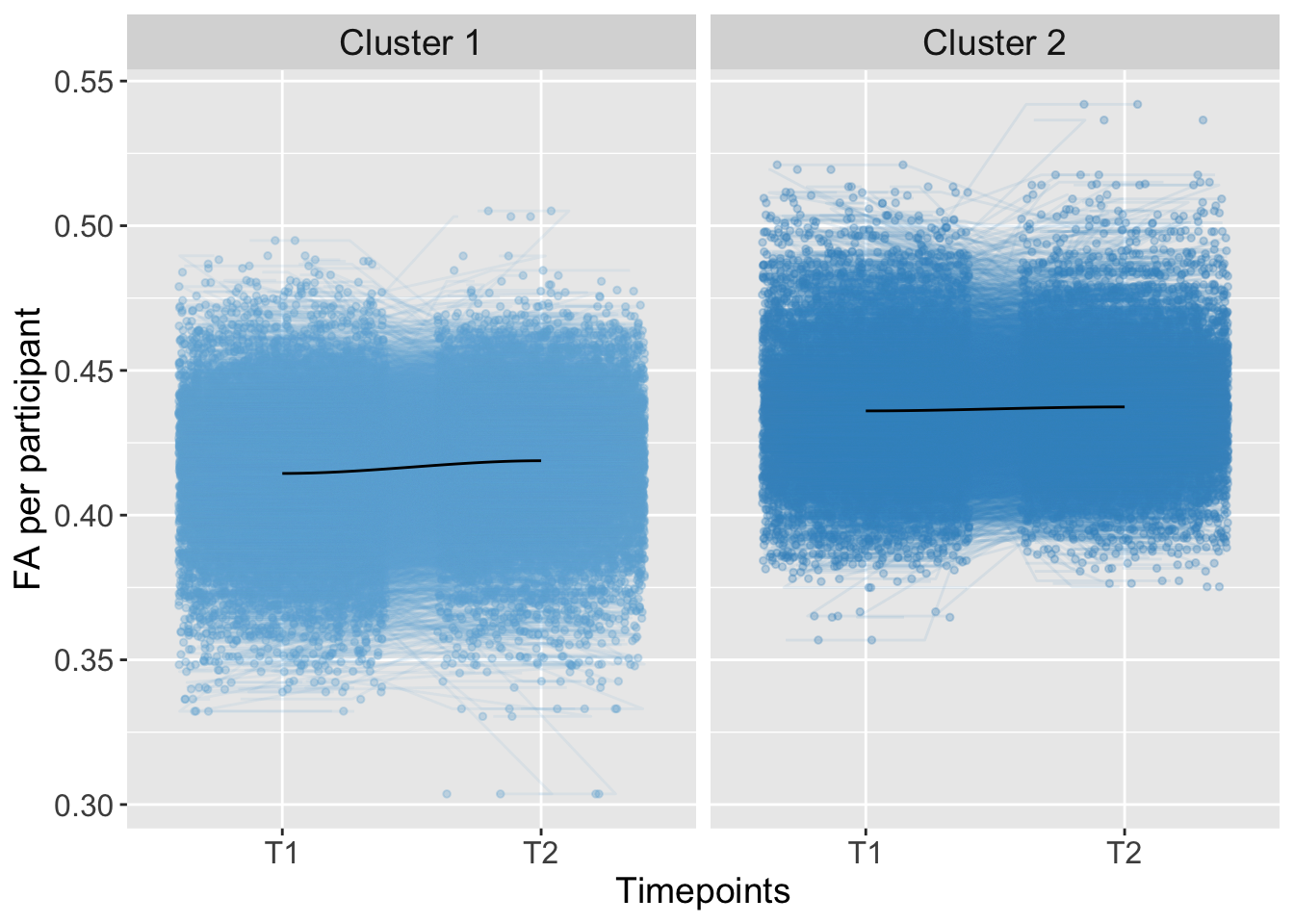
Regression results
mod_PTSD_Magnitude = multinom(MagnitudeGroups ~ PTSDSum, data=AdvDTI)
mod_School_Magnitude = multinom(MagnitudeGroups ~ SchoolSum, data=AdvDTI)
mod_Family_Magnitude = multinom(MagnitudeGroups ~ FamilySum, data=AdvDTI)
mod_Safety_Magnitude = multinom(MagnitudeGroups ~ Safety, data=AdvDTI)
mod_SES_Magnitude = multinom(MagnitudeGroups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 0.0031428 1 0.9553summary(mod_PTSD_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.165814362 0.01930412
## PTSDSum -0.001099827 0.01961957
##
## Residual Deviance: 14909.03
## AIC: 14913.03zPTMagnitude <- summary(mod_PTSD_Magnitude)$coefficients/summary(mod_PTSD_Magnitude)$standard.errors
zPTMagnitude## (Intercept) PTSDSum
## -8.58958318 -0.05605763pPTMagnitude <- (1 - pnorm(abs(zPTMagnitude), 0, 1)) * 2
pPTMagnitude## (Intercept) PTSDSum
## 0.0000000 0.9552959Anova(mod_School_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 0.04832 1 0.826summary(mod_School_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.165834520 0.01930437
## SchoolSum -0.004275047 0.01944912
##
## Residual Deviance: 14908.99
## AIC: 14912.99zSMagnitude <- summary(mod_School_Magnitude)$coefficients/summary(mod_School_Magnitude)$standard.errors
zSMagnitude## (Intercept) SchoolSum
## -8.5905150 -0.2198067pSMagnitude <- (1 - pnorm(abs(zSMagnitude), 0, 1)) * 2
pSMagnitude## (Intercept) SchoolSum
## 0.0000000 0.8260217Anova(mod_Family_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## FamilySum 21.804 1 0.00000302 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Family_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.16542719 0.01932360
## FamilySum 0.09030132 0.01934784
##
## Residual Deviance: 14887.23
## AIC: 14891.23zFMagnitude <- summary(mod_Family_Magnitude)$coefficients/summary(mod_Family_Magnitude)$standard.errors
zFMagnitude## (Intercept) FamilySum
## -8.560891 4.667255pFMagnitude <- (1 - pnorm(abs(zFMagnitude), 0, 1)) * 2
pFMagnitude## (Intercept) FamilySum
## 0.000000000000 0.000003052499Anova(mod_Safety_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## Safety 0.53333 1 0.4652summary(mod_Safety_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ Safety, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.16576385 0.01930459
## Safety 0.01418832 0.01942515
##
## Residual Deviance: 14908.5
## AIC: 14912.5zSafMagnitude <- summary(mod_Safety_Magnitude)$coefficients/summary(mod_Safety_Magnitude)$standard.errors
zSafMagnitude## (Intercept) Safety
## -8.5867572 0.7304096pSafMagnitude <- (1 - pnorm(abs(zSafMagnitude), 0, 1)) * 2
pSafMagnitude## (Intercept) Safety
## 0.0000000 0.4651399Anova(mod_SES_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## SES 1.9585 1 0.1617summary(mod_SES_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ SES, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.16552618 0.01930668
## SES 0.02721682 0.01944367
##
## Residual Deviance: 14907.08
## AIC: 14911.08zSESMagnitude <- summary(mod_SES_Magnitude)$coefficients/summary(mod_SES_Magnitude)$standard.errors
zSESMagnitude## (Intercept) SES
## -8.573519 1.399778pSESMagnitude <- (1 - pnorm(abs(zSESMagnitude), 0, 1)) * 2
pSESMagnitude## (Intercept) SES
## 0.0000000 0.1615798Ayla Pollmann - 2023