Corpus Callosum
In this script, you can find the results of the analysis across the corpus callosum. To examine individual differences, we used K-means clustering in an exploratory analysis:
K-means clustering was conducted based on the total level of brain connectivity (total; FA at imaging timepoint one plus timepoint two: higher FA total level implies greater connectivity, lower FA total levels imply lower connectivity) and the development of brain connectivity from imaging timepoint one to two (trajectory; FA at timepoint two minus timepoint one).
Multinomial logistic regression was used to predict cluster membership based on adversity experiences at baseline. To adjust for multiple comparisons, p-values were Bonferroni-corrected in the actual paper. We corrected for the number of brain regions (four) and adversity scales (five).
Total & Trajectory
K-means Clustering
set.seed(2111)
Kmeans_CC <- kmeans(AdvDTI[,19:20], 4, iter.max = 10000, nstart = 50, algorithm="Lloyd") Groups <- as.factor(Kmeans_CC$cluster)
samplesize <- 10808
id=factor(1:samplesize)
PlotCluster <- data.frame(c(AdvDTI$CC_FA_t1), #select FA at time 1
c(AdvDTI$CC_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(Groups)) #insert cluster number
colnames(PlotCluster)<-c('T1','T2','ID','Cluster') #give column name
PlotAllClusters<-melt(PlotCluster,by='ID')
ggplot(PlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2","3","4")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2", "3" = "Cluster 3", "4" = "Cluster 4")))+
scale_color_manual(values = my_colors,
breaks =c("1","2","3","4"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))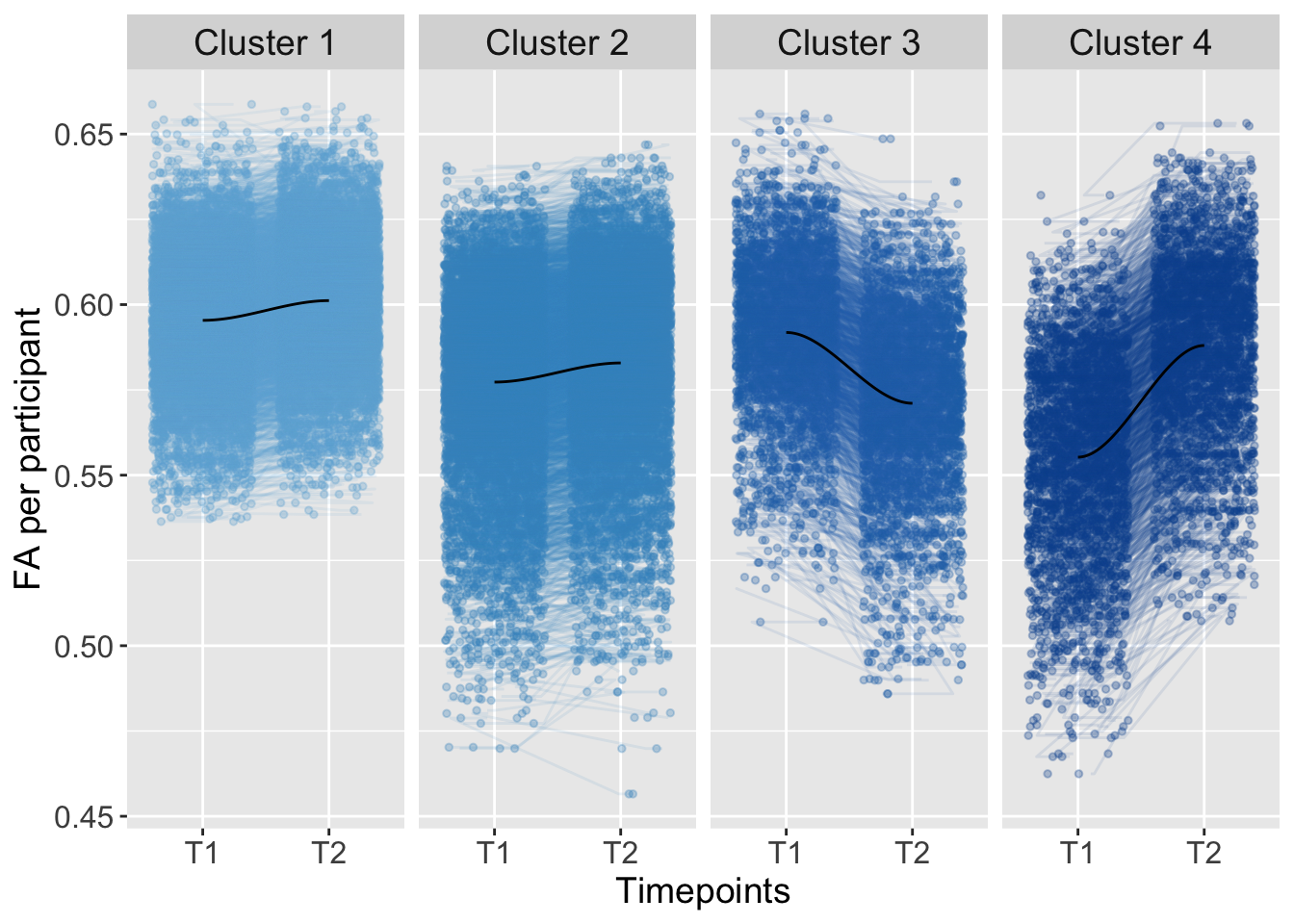
Regression results
AdvDTI$Groups <- as.factor(Kmeans_CC$cluster)
AdvDTI[,2:20] <- apply(AdvDTI[,2:20],2, function(x) as.numeric(as.character(x)))
mod_PTSD_CC = multinom(Groups ~ PTSDSum, data=AdvDTI)
mod_School_CC = multinom(Groups ~ SchoolSum, data=AdvDTI)
mod_Family_CC = multinom(Groups ~ FamilySum, data=AdvDTI)
mod_Safety_CC = multinom(Groups ~ Safety, data=AdvDTI)
mod_SES_CC = multinom(Groups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_CC, type=3) ## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 4.0034 3 0.2611summary(mod_PTSD_CC)## Call:
## multinom(formula = Groups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) PTSDSum
## 2 -0.05504257 -0.0144205561
## 3 -0.95933865 -0.0001922317
## 4 -1.14673339 0.0474860624
##
## Std. Errors:
## (Intercept) PTSDSum
## 2 0.02244705 0.02335901
## 3 0.02973852 0.03043780
## 4 0.03188719 0.02981463
##
## Residual Deviance: 27442.85
## AIC: 27454.85zPTCC <- summary(mod_PTSD_CC)$coefficients/summary(mod_PTSD_CC)$standard.errors
zPTCC## (Intercept) PTSDSum
## 2 -2.452107 -0.61734442
## 3 -32.259129 -0.00631556
## 4 -35.962194 1.59271031pPTCC <- (1 - pnorm(abs(zPTCC), 0, 1)) * 2
pPTCC## (Intercept) PTSDSum
## 2 0.01420226 0.5370076
## 3 0.00000000 0.9949609
## 4 0.00000000 0.1112252Anova(mod_School_CC, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 0.93765 3 0.8163summary(mod_School_CC)## Call:
## multinom(formula = Groups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) SchoolSum
## 2 -0.05480903 0.016004625
## 3 -0.95940772 -0.004463833
## 4 -1.14571188 0.020477267
##
## Std. Errors:
## (Intercept) SchoolSum
## 2 0.02244592 0.02260664
## 3 0.02974209 0.03008998
## 4 0.03186658 0.03199942
##
## Residual Deviance: 27445.91
## AIC: 27457.91zSCC <- summary(mod_School_CC)$coefficients/summary(mod_School_CC)$standard.errors
zSCC## (Intercept) SchoolSum
## 2 -2.441826 0.7079612
## 3 -32.257573 -0.1483494
## 4 -35.953402 0.6399263pSCC <- (1 - pnorm(abs(zSCC), 0, 1)) * 2
pSCC## (Intercept) SchoolSum
## 2 0.01461317 0.4789694
## 3 0.00000000 0.8820670
## 4 0.00000000 0.5222205Anova(mod_Family_CC, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## FamilySum 14.986 3 0.001828 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Family_CC)## Call:
## multinom(formula = Groups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## (Intercept) FamilySum
## 2 -0.0555754 -0.08578408
## 3 -0.9589762 -0.05647759
## 4 -1.1451671 -0.03078163
##
## Std. Errors:
## (Intercept) FamilySum
## 2 0.02246666 0.02257133
## 3 0.02974835 0.02982602
## 4 0.03187001 0.03168774
##
## Residual Deviance: 27431.86
## AIC: 27443.86zFCC <- summary(mod_Family_CC)$coefficients/summary(mod_Family_CC)$standard.errors
zFCC## (Intercept) FamilySum
## 2 -2.473684 -3.8005767
## 3 -32.236277 -1.8935679
## 4 -35.932435 -0.9714052pFCC <- (1 - pnorm(abs(zFCC), 0, 1)) * 2
pFCC## (Intercept) FamilySum
## 2 0.01337281 0.0001443597
## 3 0.00000000 0.0582823911
## 4 0.00000000 0.3313465187Anova(mod_Safety_CC, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## Safety 10.239 3 0.01664 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Safety_CC)## Call:
## multinom(formula = Groups ~ Safety, data = AdvDTI)
##
## Coefficients:
## (Intercept) Safety
## 2 -0.05555568 -0.06824495927
## 3 -0.95906929 -0.02812802220
## 4 -1.14576658 0.00003052823
##
## Std. Errors:
## (Intercept) Safety
## 2 0.02245926 0.02270873
## 3 0.02974083 0.02983788
## 4 0.03187434 0.03164558
##
## Residual Deviance: 27436.61
## AIC: 27448.61zSafCC <- summary(mod_Safety_CC)$coefficients/summary(mod_Safety_CC)$standard.errors
zSafCC## (Intercept) Safety
## 2 -2.473621 -3.0052294717
## 3 -32.247564 -0.9426949950
## 4 -35.946367 0.0009646918pSafCC <- (1 - pnorm(abs(zSafCC), 0, 1)) * 2
pSafCC## (Intercept) Safety
## 2 0.01337517 0.002653806
## 3 0.00000000 0.345836934
## 4 0.00000000 0.999230287Anova(mod_SES_CC, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: Groups
## LR Chisq Df Pr(>Chisq)
## SES 23.643 3 0.00002965 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_SES_CC)## Call:
## multinom(formula = Groups ~ SES, data = AdvDTI)
##
## Coefficients:
## (Intercept) SES
## 2 -0.05667253 -0.04823379
## 3 -0.96183836 0.08531301
## 4 -1.14632337 0.05027420
##
## Std. Errors:
## (Intercept) SES
## 2 0.02246837 0.02303971
## 3 0.02979329 0.02901448
## 4 0.03188172 0.03150313
##
## Residual Deviance: 27423.21
## AIC: 27435.21zSESCC <- summary(mod_SES_CC)$coefficients/summary(mod_SES_CC)$standard.errors
zSESCC## (Intercept) SES
## 2 -2.522325 -2.093507
## 3 -32.283722 2.940359
## 4 -35.955501 1.595848pSESCC <- (1 - pnorm(abs(zSESCC), 0, 1)) * 2
pSESCC## (Intercept) SES
## 2 0.0116582 0.036303911
## 3 0.0000000 0.003278318
## 4 0.0000000 0.110522771Trajectory
K-means Clustering
set.seed(2343)
Kmeans_Shape <- kmeans(AdvDTI$CC_FA_Shape, 3, iter.max = 10, nstart = 50)ShapeGroups <- as.factor(Kmeans_Shape$cluster)
ShapePlotCluster <- data.frame(c(AdvDTI$CC_FA_t1),
c(AdvDTI$CC_FA_t2),
as.factor(c(id,id)),
as.factor(ShapeGroups))
colnames(ShapePlotCluster)<-c('T1', 'T2','ID','Cluster')
ShapePlotAllClusters<-melt(ShapePlotCluster,by='ID')
ggplot(ShapePlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2","3")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2", "3" = "Cluster 3")))+
scale_color_manual(values = my_colors,
breaks =c("1","2","3"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))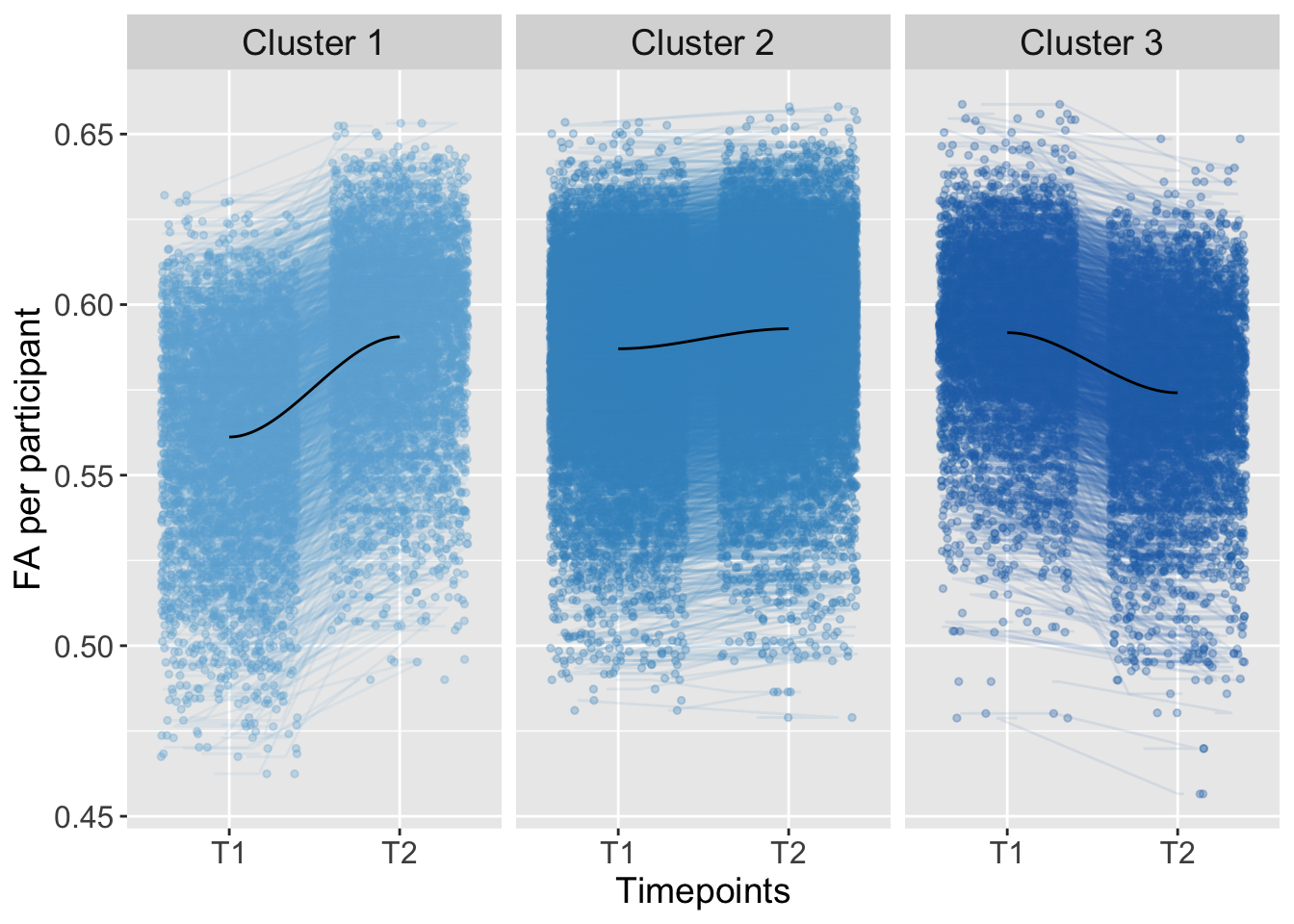
Regression results
mod_PTSD_Shape = multinom(ShapeGroups ~ PTSDSum, data=AdvDTI)
mod_School_Shape = multinom(ShapeGroups ~ SchoolSum, data=AdvDTI)
mod_Family_Shape = multinom(ShapeGroups ~ FamilySum, data=AdvDTI)
mod_Safety_Shape = multinom(ShapeGroups ~ Safety, data=AdvDTI)
mod_SES_Shape = multinom(ShapeGroups ~ SES, data=AdvDTI)Anova(mod_PTSD_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 6.0806 2 0.04782 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_PTSD_Shape)## Call:
## multinom(formula = ShapeGroups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) PTSDSum
## 2 1.295534 -0.06223648
## 3 0.171315 -0.03952417
##
## Std. Errors:
## (Intercept) PTSDSum
## 2 0.02624678 0.02493114
## 3 0.03156409 0.03015473
##
## Residual Deviance: 19871.6
## AIC: 19879.6zPTShape <- summary(mod_PTSD_Shape)$coefficients/summary(mod_PTSD_Shape)$standard.errors
zPTShape## (Intercept) PTSDSum
## 2 49.359717 -2.496335
## 3 5.427527 -1.310712pPTShape <- (1 - pnorm(abs(zPTShape), 0, 1)) * 2
pPTShape## (Intercept) PTSDSum
## 2 0.00000000000000 0.01254839
## 3 0.00000005714019 0.18995512Anova(mod_School_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 0.13255 2 0.9359summary(mod_School_Shape)## Call:
## multinom(formula = ShapeGroups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## (Intercept) SchoolSum
## 2 1.294815 0.008551055
## 3 0.170455 0.010780702
##
## Std. Errors:
## (Intercept) SchoolSum
## 2 0.02622922 0.02648226
## 3 0.03155011 0.03181533
##
## Residual Deviance: 19877.55
## AIC: 19885.55zSShape <- summary(mod_School_Shape)$coefficients/summary(mod_School_Shape)$standard.errors
zSShape## (Intercept) SchoolSum
## 2 49.365374 0.3228975
## 3 5.402677 0.3388524pSShape <- (1 - pnorm(abs(zSShape), 0, 1)) * 2
pSShape## (Intercept) SchoolSum
## 2 0.00000000000000 0.7467729
## 3 0.00000006565352 0.7347209Anova(mod_Family_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## FamilySum 0.4241 2 0.8089summary(mod_Family_Shape)## Call:
## multinom(formula = ShapeGroups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## (Intercept) FamilySum
## 2 1.2947374 -0.006155084
## 3 0.1701906 -0.019427289
##
## Std. Errors:
## (Intercept) FamilySum
## 2 0.02622765 0.02621389
## 3 0.03155144 0.03164674
##
## Residual Deviance: 19877.26
## AIC: 19885.26zFShape <- summary(mod_Family_Shape)$coefficients/summary(mod_Family_Shape)$standard.errors
zFShape## (Intercept) FamilySum
## 2 49.365355 -0.2348024
## 3 5.394068 -0.6138796pFShape <- (1 - pnorm(abs(zFShape), 0, 1)) * 2
pFShape## (Intercept) FamilySum
## 2 0.00000000000000 0.8143621
## 3 0.00000006888014 0.5392949Anova(mod_Safety_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## Safety 0.8205 2 0.6635summary(mod_Safety_Shape)## Call:
## multinom(formula = ShapeGroups ~ Safety, data = AdvDTI)
##
## Coefficients:
## (Intercept) Safety
## 2 1.2947483 -0.02031187
## 3 0.1704031 -0.00450179
##
## Std. Errors:
## (Intercept) Safety
## 2 0.02622885 0.02628586
## 3 0.03154985 0.03158046
##
## Residual Deviance: 19876.86
## AIC: 19884.86zSafShape <- summary(mod_Safety_Shape)$coefficients/summary(mod_Safety_Shape)$standard.errors
zSafShape## (Intercept) Safety
## 2 49.363523 -0.7727300
## 3 5.401074 -0.1425499pSafShape <- (1 - pnorm(abs(zSafShape), 0, 1)) * 2
pSafShape## (Intercept) Safety
## 2 0.00000000000000 0.4396822
## 3 0.00000006624316 0.8866457Anova(mod_SES_Shape, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: ShapeGroups
## LR Chisq Df Pr(>Chisq)
## SES 22.509 2 0.00001295 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_SES_Shape)## Call:
## multinom(formula = ShapeGroups ~ SES, data = AdvDTI)
##
## Coefficients:
## (Intercept) SES
## 2 1.2937990 -0.05426966
## 3 0.1680632 0.05865348
##
## Std. Errors:
## (Intercept) SES
## 2 0.02623414 0.02634861
## 3 0.03158253 0.03097238
##
## Residual Deviance: 19855.17
## AIC: 19863.17zSESShape <- summary(mod_SES_Shape)$coefficients/summary(mod_SES_Shape)$standard.errors
zSESShape## (Intercept) SES
## 2 49.317387 -2.059678
## 3 5.321397 1.893735pSESShape <- (1 - pnorm(abs(zSESShape), 0, 1)) * 2
pSESShape## (Intercept) SES
## 2 0.0000000000000 0.03942933
## 3 0.0000001029736 0.05826018Total
K-means Clustering
set.seed(123)
Kmeans_Magnitude <- kmeans(AdvDTI[,20], 2, iter.max = 10, nstart = 50) #clusterMagnitudeGroups <- as.factor(Kmeans_Magnitude$cluster)
MagnitudePlotCluster <- data.frame(c(AdvDTI$CC_FA_t1), #select FA at time 1
c(AdvDTI$CC_FA_t2), #select FA at time 2
as.factor(c(id,id)), #Give ID to participants
as.factor(MagnitudeGroups)) #insert cluster number
colnames(MagnitudePlotCluster)<-c('T1', 'T2','ID','Cluster') #give column name
MagnitudePlotAllClusters<-melt(MagnitudePlotCluster,by='ID')
ggplot(MagnitudePlotAllClusters,aes(variable,value,group=ID,col=Cluster))+
geom_point(position = "jitter", size=1,alpha=.3)+
geom_line(position = "jitter", alpha=.1)+
stat_smooth(aes(group=Cluster), colour="black", method="loess", se=F, size=0.5, formula = 'y~x')+
ylab('FA per participant')+
xlab('Timepoints')+
facet_grid(~factor(Cluster, levels=c("1","2")),
labeller = as_labeller(c("1" = "Cluster 1", "2" = "Cluster 2")))+
scale_color_manual(values = my_colors,
breaks =c("1","2","3","4"))+
guides(col = FALSE)+
theme(axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
strip.text = element_text(size = 14))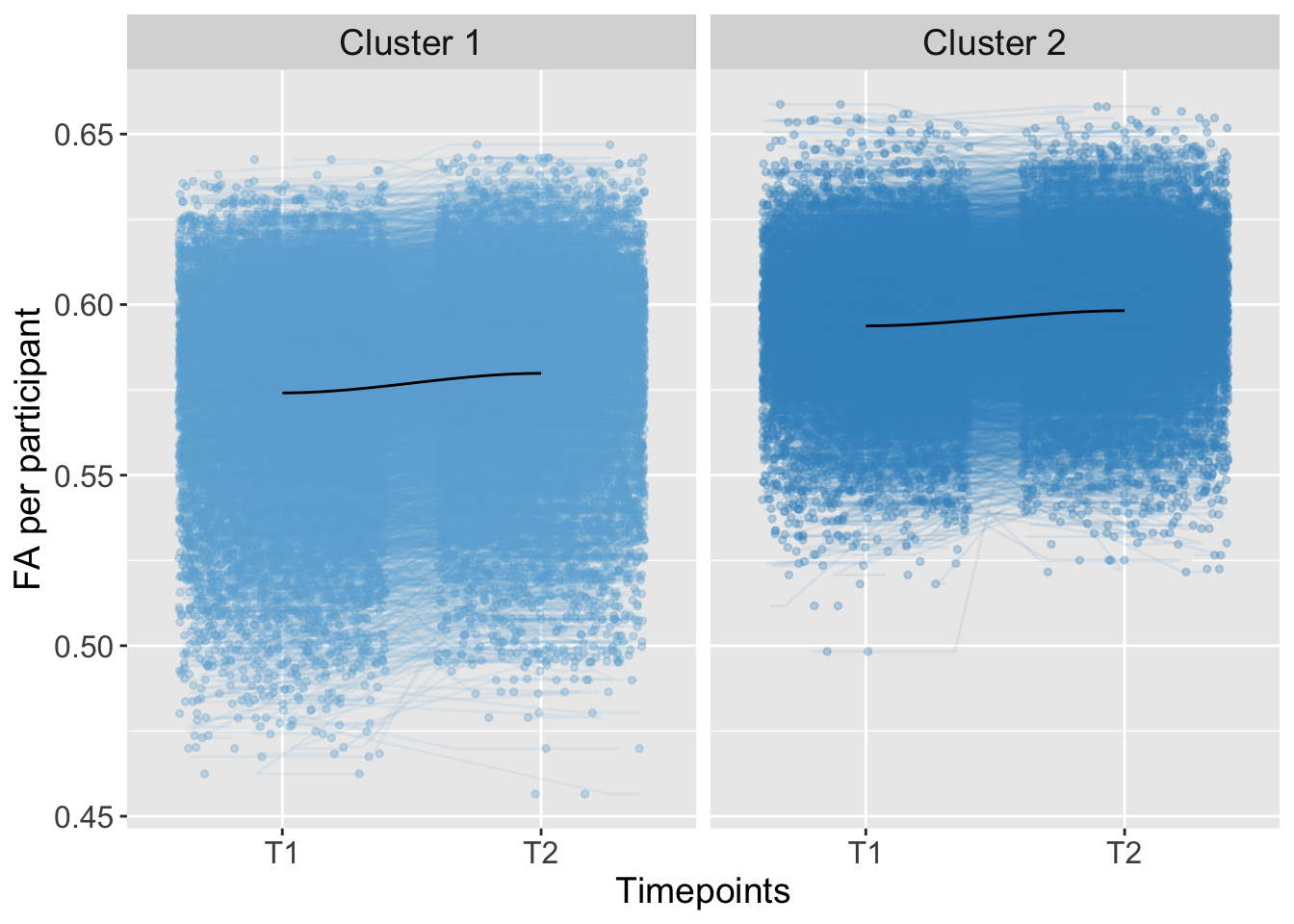
Regression results
mod_PTSD_Magnitude = multinom(MagnitudeGroups ~ PTSDSum, data=AdvDTI)
mod_School_Magnitude = multinom(MagnitudeGroups ~ SchoolSum, data=AdvDTI)
mod_Family_Magnitude = multinom(MagnitudeGroups ~ FamilySum, data=AdvDTI)
mod_Safety_Magnitude = multinom(MagnitudeGroups ~ Safety, data=AdvDTI)
mod_SES_Magnitude = multinom(MagnitudeGroups ~ SES, data=AdvDTI)options(scipen = 999)
Anova(mod_PTSD_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## PTSDSum 0.15745 1 0.6915summary(mod_PTSD_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ PTSDSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.071438277 0.01925037
## PTSDSum 0.007759964 0.01955630
##
## Residual Deviance: 14969.12
## AIC: 14973.12zPTMagnitude <- summary(mod_PTSD_Magnitude)$coefficients/summary(mod_PTSD_Magnitude)$standard.errors
zPTMagnitude## (Intercept) PTSDSum
## -3.7110076 0.3968013pPTMagnitude <- (1 - pnorm(abs(zPTMagnitude), 0, 1)) * 2
pPTMagnitude## (Intercept) PTSDSum
## 0.0002064359 0.6915139609Anova(mod_School_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## SchoolSum 1.3067 1 0.253summary(mod_School_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ SchoolSum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.07158195 0.01925168
## SchoolSum -0.02217396 0.01940262
##
## Residual Deviance: 14967.97
## AIC: 14971.97zSMagnitude <- summary(mod_School_Magnitude)$coefficients/summary(mod_School_Magnitude)$standard.errors
zSMagnitude## (Intercept) SchoolSum
## -3.718218 -1.142833pSMagnitude <- (1 - pnorm(abs(zSMagnitude), 0, 1)) * 2
pSMagnitude## (Intercept) SchoolSum
## 0.0002006329 0.2531077353Anova(mod_Family_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## FamilySum 17.948 1 0.0000227 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Family_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ FamilySum, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.07093424 0.01926652
## FamilySum 0.08179353 0.01932391
##
## Residual Deviance: 14951.33
## AIC: 14955.33zFMagnitude <- summary(mod_Family_Magnitude)$coefficients/summary(mod_Family_Magnitude)$standard.errors
zFMagnitude## (Intercept) FamilySum
## -3.681736 4.232763pFMagnitude <- (1 - pnorm(abs(zFMagnitude), 0, 1)) * 2
pFMagnitude## (Intercept) FamilySum
## 0.00023165105 0.00002308377Anova(mod_Safety_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## Safety 3.0548 1 0.0805 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod_Safety_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ Safety, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.07134222 0.01925298
## Safety 0.03386783 0.01937806
##
## Residual Deviance: 14966.23
## AIC: 14970.23zSafMagnitude <- summary(mod_Safety_Magnitude)$coefficients/summary(mod_Safety_Magnitude)$standard.errors
zSafMagnitude## (Intercept) Safety
## -3.705516 1.747741pSafMagnitude <- (1 - pnorm(abs(zSafMagnitude), 0, 1)) * 2
pSafMagnitude## (Intercept) Safety
## 0.0002109607 0.0805089454Anova(mod_SES_Magnitude, type=3)## Analysis of Deviance Table (Type III tests)
##
## Response: MagnitudeGroups
## LR Chisq Df Pr(>Chisq)
## SES 2.2787 1 0.1312summary(mod_SES_Magnitude)## Call:
## multinom(formula = MagnitudeGroups ~ SES, data = AdvDTI)
##
## Coefficients:
## Values Std. Err.
## (Intercept) -0.07113386 0.01925333
## SES 0.02929034 0.01940476
##
## Residual Deviance: 14967
## AIC: 14971zSESMagnitude <- summary(mod_SES_Magnitude)$coefficients/summary(mod_SES_Magnitude)$standard.errors
zSESMagnitude## (Intercept) SES
## -3.694627 1.509441pSESMagnitude <- (1 - pnorm(abs(zSESMagnitude), 0, 1)) * 2
pSESMagnitude## (Intercept) SES
## 0.0002202099 0.1311860336Ayla Pollmann - 2023